Monthly Archives: November 2016
Epilepsie
L’épilepsie, aussi appelée mal comitial, est une affection neurologique définie depuis 2005 par la ligue internationale contre l’épilepsie (ILAE) par une prédisposition cérébrale à générer des crises épileptiques dites « non provoquées », c’est-à-dire non expliquées par un facteur causal immédiat. Une crise épileptique est caractérisée par une altération fonctionnelle transitoire au sein d’une population de neurones (soit limitée à une région du cerveau : crise dite « partielle », soit impliquant les deux hémisphères cérébraux de manière simultanée : crise dite « généralisée ») due à une décharge épileptique.
Un facteur prédisposant peut être d’origine génétique, lésionnel (lésion cérébrale présente depuis la naissance, malformative, ou acquise au cours de la vie, comme un accident vasculaire cérébral ou les complications d’un traumatisme crânien grave), ou autre (par exemple, une anomalie de l’électro-encéphalogramme peut être considérée comme prédisposant).
Il n’existe pas une seule épilepsie, mais de nombreuses formes différentes, à l’origine de crises épileptiques elles-mêmes très variées (crise tonico-clonique généralisée ou convulsions, crise myoclonique, absence épileptique, crise partielle simple, crise partielle complexe, sont les plus fréquentes). Une épilepsie est caractérisée par deux traits essentiels : le caractère « généralisé » (les crises intéressent d’emblée les deux hémisphères cérébraux) ou « partiel » (les crises n’intéressent qu’une population limitée de neurones), et leur étiologie (« idiopathique » ou « génétique », et « symptomatique » ou « structurel/métabolique », selon les anciennes et nouvelles terminologies, respectivement).
Chez les personnes souffrant d’épilepsie, la survenue d’une crise est souvent due à un état de fatigue inhabituel, un manque de sommeil, ou un état anxieux. Dans une minorité de cas dits « photosensibles », une stimulation lumineuse intermittente (par exemple, un stroboscope) peut être à l’origine des crises.
Causes
Il existe une classification internationale des épilepsies (en). Les causes de l’épilepsie sont très nombreuses, tout le monde peut être concerné par l’apparition d’une crise d’épilepsie sans pour autant « être épileptique ». Un certain nombre d’affections diminuent la résistance à lutter contre la propagation de la décharge électrique dans le cerveau. Une baisse du seuil épileptogène serait responsable de la crise d’épilepsie.
Des crises occasionnelles, survenant dans des conditions particulières, peuvent être causées notamment par convulsions fébriles, hypoglycémie, troubles ioniques, hypocalcémie, hyponatrémie, alcoolisme, ivresse aiguë, sevrage alcoolique, surdosage en médicaments (théophylline) ou sevrage en médicament antiépileptique (benzodiazépines).
Les épilepsies symptomatiques peuvent être causées par tumeur cérébrale, accident vasculaire cérébral, encéphalite, traumatisme crânien et la maladie d’Alzheimer (il s’agit d’une complication méconnue, mais à la fois non systématique et à un stade avancé de cette pathologie4). Dans le cas du traumatisme crânien, une contusion cérébrale avec perte de connaissance après traumatisme, sans lésion cérébrale démontrée, double le risque de survenue d’une épilepsie. Ce risque reste majoré plus de 10 ans après l’accident initial.
D’autres types d’épilepsies incluent l’épilepsie cryptogénique (voir les syndromes épileptiques ci-dessus) dont une cause organique supposée existe, mais qui ne peut être mise en évidence, l’épilepsie idiopathique (aucune cause décelée ni suspectée), la chorée de Huntington (qui se présente surtout chez des cas jeunes) et l’épilepsie dans le contexte du syndrome du chromosome 20 en anneau.
Il existe des formes familiales d’épilepsie faisant suspecter une cause génétique, mais dont l’étude n’est pas aisée : une crise convulsive chez un individu peut survenir pour de multiples raisons, et toutes les convulsions ne sont pas dues à une maladie épileptique mais au franchissement du seuil épileptogène ; les maladies épileptiques n’étant que des affections ayant pour corollaire la baisse de ce seuil à un niveau anormalement bas. Ce seuil est facilement franchissable lors de petites provocations rencontrées dans la vie quotidienne (jeûne, alcool, fatigue, drogues fortes…).
Il semble que les maladies auto-immunes soient un facteur prédisposant à l’épilepsie.
Il semble que l’exposition prénatale à l’alcool augmente le risque d’épilepsie.
La convulsion fébrile en pédiatrie peut également mener au développement de l’épilepsie, mais cette cause est plutôt rare. En effet, environ 1 enfant sur 40 va développer de l’épilepsie suite aux épisodes de convulsions fébriles en pédiatrie.
Diagnostic – Signes et symptômes
L’épilepsie est un symptôme neurologique causé par un dysfonctionnement passager du cerveau ; certains disent qu’il « court-circuite ». Lors d’une crise d’épilepsie, les neurones (cellules nerveuses cérébrales) produisent soudainement une décharge électrique anormale dans certaines zones cérébrales.
Bien que l’épilepsie touche un grand nombre de personnes par ailleurs normales, le prévalence de ce trouble est fréquente chez les personnes atteintes d’arriération mentale. Réciproquement, 30% des enfants atteints d’épilepsie ont également un retard de développement. Chez l’enfant, une blessure au cerveau après la naissance est associé à une forte prévalence de l’épilepsie (75%).
Tout le monde peut être concerné par une première crise d’épilepsie, mais dans la moitié des cas il n’y en aura plus jamais d’autres. Il n’était jusque-là question d’épilepsie que si les crises se répétaient, mais depuis 2006 les médecins s’accordent à dire qu’une seule suffit pour être épileptique.
Chez l’enfant
Chez les enfants, l’épilepsie disparaît à l’adolescence dans six cas sur 10, lorsque les circuits neuronaux ont fini leur développement. Dans deux autres cas sur 10, les médicaments peuvent être supprimés après plusieurs années de traitement sans crise.
Le valium et le phénobarbital stoppent dans un premier temps les crises épileptiques de l’enfant, mais peuvent les aggraver après plusieurs traitements. Ces médicaments renforcent en effet l’action du GABA10, médiateur de l’inhibition cérébrale, qui permet en temps normal l’entrée d’ions chlorure (négatifs) dans le neurone, en se fixant sur son récepteur GABAR (molécule–canal). Or, il est montré chez le rat de laboratoire que le GABA – après plusieurs crises – est responsable d’un excès d’accumulation d’ions chlorure dans les neurones11. Le GABA cause alors une sortie excessive d’ions chlorure (sortie renforcée par le phénobarbital). De plus, au fur et à mesure des crises, le transporteur KCC2 – expulsant des ions chlorure – fonctionne moins bien, alors que NKCC1 – un autre cotransporteur expulsant des ions chlorure – continue à fonctionner12. Un diurétique précocement prescrit avec le phénobarbital, en freinant l’activité de KNCC1 du rein, permettrait de diminuer le risque de crise.
Crises généralisées
La crise tonico-clonique, ou « grand mal », représente la forme la plus spectaculaire de crise d’épilepsie. Le patient perd connaissance brutalement et son organisme présente des manifestations évoluant en trois phases : phase tonique, causant raidissement, contraction de l’ensemble des muscles des membres, du tronc et du visage dont les muscles oculomoteurs et masticateurs ; phase clonique causant des convulsions, contractions désordonnées des mêmes muscles et récupération, phase stertoreuse (perte de connaissance se prolongeant durant quelques minutes à quelques heures), caractérisée par une respiration bruyante due à l’encombrement bronchique. Cette phase est une phase de relaxation intense durant laquelle il est possible mais pas systématique de perdre ses urines. Le retour à la conscience est progressif, il existe souvent une confusion post-critique et l’absence de souvenir de la crise.
Les absences représentent une forme fréquente d’épilepsie et concernent quasiment exclusivement les enfants (jusqu’à la puberté). Elles se manifestent par une perte brusque du contact avec regard vitreux, aréactivité aux stimuli, des phénomènes cloniques (clignement des paupières, spasmes de la face, mâchonnements), parfois toniques (raidissement du tronc) ou végétatifs (perte d’urines, hypersalivation). Dans les absences typiques, la perte de conscience et le retour à la conscience sont brutaux, l’absence durant quelques secondes. Les chutes au sol sont rares. Dans certains cas, l’absence n’est ni précédée de symptômes précurseurs, ni suivie d’un ressenti particulier. Aussi, sans témoignage extérieur, la personne épileptique n’a parfois aucun moyen de savoir qu’elle a fait une crise. Elles sont contemporaines d’une activité EEG caractéristique : pointes-ondes de 3Hz, bilatérales, symétriques et synchrones.
Les absences peuvent se répéter dix à cent fois par jour en l’absence de traitement.
Les myoclonies, également une cause, se manifestent par des secousses musculaires brutales, rythmées, intenses, bilatérales ou unilatérales et synchrones, concernant les bras ou les jambes, sans perte de la conscience mais occasionnant des chutes au sol.
Le diagnostic n’est cependant pas toujours évident. Ainsi une syncope d’origine cardiaque peut se manifester, outre la perte de connaissance, par des mouvements cloniques. Le moniteur cardiaque implantable peut ainsi redresser un certain nombre de diagnostic erroné d’épilepsie qui sont « guéris » par la pose d’un stimulateur cardiaque.
Crises partielles
La symptomatologie est extrêmement polymorphe (il peut y avoir de nombreux symptômes) : le foyer épileptique est circonscrit à une zone limitée du cerveau, et entraîne des signes cliniques corrélés à la zone touchée. Les signes peuvent être l’un des items suivant :
- des hallucinations sensorielles (visuelles, auditives, gustatives, vertigineuses)
- des mouvements anormaux (crise tonico-clonique focale) ou une paralysie des muscles d’un membre, de la tête, de la voix…
- des troubles de la sensibilité (engourdissement, paresthésies)
- des troubles d’apparence psychiatrique tels qu’une dysmnésie (flashbacks ou ecmnésie), des hallucinations psychotiques, une métamorphopsie (sensation de distorsion des objets) et des angoisses phobiques
- des troubles du langage sur le versant expressif et/ou sur le versant réceptif : modifications du débit de parole, perte de l’intelligibilité, trouble de la compréhension…
- des troubles du système nerveux végétatif :
- tachycardie, hypotension, vomissements, diarrhées, dyspepsie (indigestion), hyper-salivation, gastralgies, reflux gastro-œsophagien.
Les crises partielles simples ne s’accompagnent pas de troubles de la conscience, à la différence des crises partielles complexes. Dans certains cas, les crises partielles peuvent se généraliser (tonico-cloniques) dans un second temps par extension à tout l’encéphale de la crise épileptique.
Syndromes épileptiques
Un certain nombre de syndromes épileptiques ont été individualisés, caractérisés par le contexte clinique, les symptômes épileptiques, et les résultats de l’électro-encéphalogramme.
- Syndrome de West : affection grave touchant le nourrisson avant un an. Caractérisé par des spasmes, des troubles psychomoteurs avec mauvais développement intellectuel, et un électroencéphalogramme montrant une hypsarythmie typique. L’enfant gardera des séquelles neurologiques sévères, avec un bas pourcentage d’exception.
- Syndrome de Lennox-Gastaut : affection grave touchant les jeunes enfants de deux à six ans. Des crises généralisées toniques ou des absences pluri-quotidiennes, des troubles intellectuels sont diagnostiqués. L’électroencéphalogramme entre deux crises est également typique. L’enfant gardera habituellement des séquelles neurologiques plus ou moins sévères. Cependant des cas rares peuvent évoluer sans séquelles.
- Syndrome de Dravet, longtemps appelé « épilepsie myoclonique sévère du nourrisson » ou EMSN, comporte des crises convulsives sensibles à la fièvre qui peuvent être généralisées ou partielles. Il débute souvent entre 4 et 8 mois chez un nourrisson sans antécédent ni retard préexistant mais peut se déclencher plus tard, et qui n’accusera un retard que dans le courant de la deuxième année. L’épilepsie débute par des crises convulsives, unilatérales ou généralisées, spontanées ou provoquées par la fièvre, voire par un vaccin ; l’évolution est différente selon les cas mais souvent caractérisée par une instabilité du comportement, de la motricité, et un retard du langage. (source epilepsie France)
- Épilepsie d’absence de l’enfant
- Épilepsie myoclonique juvénile : maladie apparaissant à l’adolescence faite de crises myocloniques, avec un électro-encéphalogramme anormal. Évolution favorable. Elle se caractérise par des spasmes musculaires non contrôlés dont les signaux électriques du cerveau dure (généralement) quelques millièmes de seconde. Apparaissent, lors d’une crise longue de quelques minutes, certaines pertes sensorielles, dont l’impossibilité à trouver des mots permettant d’exprimer la pensée, ainsi que l’absence des sens spatio-temporels. À ce jour, aucun médicament ne peut arrêter formellement une crise épileptique myoclonique (juvénile ou non), mais quelques traitements permettent aujourd’hui l’atténuation des crises au fur et à mesure des années.
- Épilepsie frontale à crises nocturnes
- Convulsions fébriles de l’enfant : ces convulsions sont généralisées (tonico-cloniques) et apparaissent chez l’enfant, entre six mois et cinq ans, à l’occasion d’une hyperthermie, quelle qu’en soit l’origine — sauf pour la méningite qui est la cause directe des crises convulsives. Ces enfants ont une sensibilité supérieure aux autres enfants à faire des convulsions dans un contexte fébrile (lors d’une fièvre). Cette affection disparaît avec l’âge. L’électro-encéphalogramme est quasi-normal entre les crises.
Données anatomiques et fonctionnelles
Les données anatomiques permettent de mettre en évidence l’existence ou non de structures cérébrales endommagées et donc de connaître le foyer lésionnel susceptible de générer les crises d’épilepsie. Ces données sont enregistrées grâce à plusieurs modalités d’imagerie cérébrale telles que les images radiologiques, les scanners ou encore les Imagerie par Résonance Magnétique (IRM). De plus grâce aux progrès faits dans le domaine de l’imagerie encéphalique ces dernières années, il est aujourd’hui possible d’enregistrer les zones de fonctionnement de certaines parties du cerveau grâce à l’IRMf.
Modèles animaux de l’épilepsie
Étant donné que l’épilepsie semble être plus un groupe de symptômes qu’une maladie unique, plusieurs modèles sont utilisés pour étudier les mécanismes biologiques de l’épilepsie.
- Une crise convulsive peut être provoquée chez de jeunes rats ou de jeunes souris (10 à 11 jours après la naissance) par une hyperthermie pendant 30 minutes afin de modéliser une crise fébrile. Chez certains rats (environ 30%), des crises focales peuvent être observées lorsqu’ils sont adultes.
- Un état de mal épileptique (status epilepticus) peut être provoqué par l’injection intrapéritonéale de pilocarpine, un agoniste des récepteurs muscariniques ou celle de kaïnate, un agoniste des récepteurs glutamatergiques. Après un tel traitement, l’animal développe un profil EEG comparable à celui d’un état de mal épileptique chez l’homme et doit souvent être interrompu par l’injection d’une benzodiazépine (valium). Plusieurs semaines après cet état de mal, les animaux présentent des crises convulsives spontanées récurrente et des lésions dans l’hippocampe et d’autres structures limbiques qui évoquent une sclérose de l’hippocampe. Ces préparations, ou celles qui sont obtenues par l’injection dans l’hippocampe de kaïnate sont utilisées comme modèles d’épilepsie mésiotemporale.
- Il existe aussi des animaux (rats, souris) qui possèdent une ou plusieurs mutations génétiques qui les rend épileptiques. En particulier, les GAERS (genetic absence epilepsy rat from Strasbourg) découverts à Strasbourg en 1982, présentent des crises d’épilepsie spontanées, non-convulsives caractérisées par un arrêt du comportement et la survenue de décharges de pointe-onde sur l’EEG. Ces crises et leur réactivité à certains antiépileptiques (valproate, éthosuximide) font du GAERS un modèle d’épilepsie absence 19. Une lignée de rats sans aucune crise a également été sélectionnée (NEC : Non Epileptic Control) à partir des mêmes animaux de départ (Wistar).
Corrélats biologiques
Pendant une crise épileptique, un grand nombre de neurones déchargent des potentiels d’action de manière synchronisée. La synchronisation des neurones est une propriété intrinsèque des réseaux neuronaux. L’observation d’EEG de sujets sains révèle l’existence de différentes fréquences propres à l’activité cérébrale qui correspondent à des synchronisations de groupes de neurones. Il semblerait que, dans le cas de l’épilepsie, cette synchronisation s’emballerait. Plus exactement, un dysfonctionnement se passe dans la transmission synaptique. Lors de cette transmission, des neurotransmetteurs ne partent pas de la terminaison pré-synaptique à la terminaison cible post-synaptique mais certains se rattachent à d’autres synapses. Ce phénomène est de l’ordre de 5 % à la normale, mais dans les épilepsies généralisées, il est de 25 %.
Plusieurs causes peuvent expliquer cet « emballement » :
- une surexcitabilité des neurones ;
- une diminution du niveau d’inhibition du réseau neuronal.
Une crise épileptique provoque sur chaque neurone concerné une série de changements qui peuvent expliquer qu’une épilepsie devienne chronique. Ces changements sont assimilables à la plasticité neuronale, laquelle explique aussi les processus de mémorisation et d’apprentissage.
L’inhibition neuronale semble contrôlée par une enzyme du métabolisme énergétique. Chez les épileptiques, ce contrôle pourrait être déficient, favorisant ainsi le déclenchement des crises.
Implication de la génétique
Il est vraisemblable qu’une multitude de gènes — dont beaucoup sont encore à identifier — interagissent de manière très complexe. Dans certaines formes génétiques familiales — concernant à peine 1 % des cas — des chercheurs australiens de l’Epilepsy Research Center de Melbourne (Australie) ont réussi à identifier une dizaine de gènes. Les recherches génétiques, qui se situent aux confins de la recherche et de la clinique, n’en sont qu’à leurs balbutiements, mais devraient pouvoir aider à l’avenir dans le diagnostic et le choix de certaines stratégies de traitements. Par exemple, dans le cas de certains types d’épilepsie, susceptibles de se compliquer d’une encéphalopathie chez de très jeunes enfants, les épileptologues espèrent pouvoir les diagnostiquer beaucoup plus tôt, dès les premiers mois de vie, grâce à la génétique moléculaire. Ceci, afin de pouvoir proposer un traitement capable de réduire la gravité de ces encéphalopathies, voire d’en empêcher la survenue.
En France, un centre de génétique moléculaire spécialement affecté à ces maladies rares a été mis en place à la Pitié-Salpêtrière.
Risques
L’épilepsie et les crises d’épilepsie ne mettent pas, en général, la vie de la personne épileptique en danger. Cependant, il est utile de mentionner quelques risques liés à l’épilepsie. La répétition des crises d’épilepsie peut entraîner l’apparition de lésions cérébrales. Ces lésions prédominent dans les régions cérébrales à l’origine des crises mais aussi dans une région particulière, la face interne du lobe temporal. Elles sont susceptibles par la suite d’être à l’origine de nouvelles crises — il est dit que la maladie s’auto-aggrave.
Lors d’une crise, le patient n’avale jamais sa langue, il est inutile (et dangereux) d’essayer de la retenir chez un patient en pleine convulsion.
Les risques liés à l’épilepsie ne concernent généralement pas la crise en elle-même, mais ses conséquences. Ainsi, si le patient conduit ou fait une mauvaise chute, à titre d’exemple, les conséquences de la crise peuvent alors être mortelles.
État de mal
L’état de mal épileptique engage le pronostic vital et fonctionnel (risque de séquelles neurologiques définitives). Elle est définie par une crise épileptique de durée anormalement longue (plus de 30 minutes), ou par la survenue de crises si rapprochées que le malade n’a pas le temps de recouvrer ses esprits entre deux crises. L’état de mal épileptique doit faire rechercher une cause déclenchante : arrêt du traitement, prise de toxiques (ivresse alcoolique en particulier), prise de médicaments contre-indiqués, infection, etc. Dans le cas d’un état de mal par crises convulsives tonico-cloniques, la durée de la crise fait craindre : insuffisance respiratoire par encombrement bronchique et bradypnée (ralentissement respiratoire), troubles circulatoires, séquelles neurologiques et intellectuelles définitives, troubles hydro-électrolytiques (déshydratation, acidose) et œdème cérébral.
L’état de mal peut aussi se produire dans le cadre du petit mal : un état confusionnel prolongé est alors retrouvé, associé à des manifestations cloniques du visage très évocatrices. En cas d’état de mal par épilepsie partielle, le diagnostic peut être difficile en raison de la ressemblance des symptômes avec ceux d’un accident vasculaire cérébral. L’électro-encéphalogramme permet de faire la différence. Dans les cas extrêmes, la crise peut conduire à la mort du patient, par anoxie cérébrale.
Spondylolisthésis par lyse isthmique
Le spondylolisthésis désigne une affection du squelette humain, caractérisée par le glissement d’une vertèbre en avant de la vertèbre située en dessous d’elle (antéspondylolisthésis) ou en arrière (rétrolisthésis).
Les vertèbres les plus souvent concernées sont la quatrième et la cinquième vertèbre lombaire.
Spondylolyse et spondylolisthésis isthmique chez l’adulte
La spondylolyse est une perte de continuité de l’isthme articulaire (pars interarticularis), situé entre les deux apophyses articulaires supérieure et inférieure de l’arc vertébral postérieur. Elle survient le plus souvent au niveau d’un étage vertébral lombaire bas, habituellement le dernier étage mobile. Elle peut être unilatérale ou bilatérale, induisant alors une micro mobilité ou une mobilité de l’arc postérieur. Elle peut être aiguë, traumatique, correspondant à un trait de fracture vrai, ou chronique, sans notion de vrai traumatisme. La perte de continuité s’organise alors sous la forme d’un tissu fibreux ou plus rarement pseudo-kystique plus ou moins hypertrophique dit « nodule de Gill », qui peut être agressif pour les racines nerveuses au contact. La lyse peut survenir dans l’enfance, mais aussi à l’âge adulte.
La fréquence de la spondylolyse au niveau lombaire est remarquablement élevée : de l’ordre de 3 à 7 % dans la population générale, beaucoup plus dans certains groupes. Si Inuits et Bushmen sont classiquement des groupes à risque, la fréquence des spondylolyses est de 14 % chez les athlètes de haut niveau, particulièrement 20 % chez les danseurs, et de 11 à 17 % chez les gymnastes et les rameurs, au point que la constatation d’une spondylolyse puisse être considérée comme presque physiologique dans certains groupes de sportifs…
Le spondylolisthésis est un déplacement permanent du corps vertébral en avant sur le corps vertébral sous-jacent, constituant un antéspondylolisthésis, ou en arrière (rétrolisthésis). Il est deux à trois fois moins fréquent que la spondylolyse isthmique. Dans sa forme dite isthmique, il survient à la faveur d’une spondylolyse vraie, ou d’un allongement « dysplasique » de l’isthme sans perte de continuité qui est peut-être le résultat de lésions traumatiques consolidées. Les spondylolisthésis ont été classés de façon purement descriptive par Newman en 5 types essentiels.
- Le type isthmique est le type II, avec une forme lytique IIA, une forme avec allongement de l’isthme sans lyse IIB, et une forme traumatique aiguë avec fracture des isthmes IIC.
- Le type I « dysplasique » et plutôt rare recouvre un ensemble de malformations congénitales de la charnière lombo-sacrée : incompétence des articulaires postérieures dysplasiques associée à une anomalie de croissance du sacrum « en dôme », il se voit chez l’enfant et s’accompagne de troubles radiculaires car l’arc postérieur est habituellement intact.
- Le type III correspond au spondylolisthésis dit « dégénératif » : il s’agit d’un glissement habituellement à l’avant dernier niveau mobile, dû à la faillite arthrosique des apophyses articulaires postérieures et permise par la dégénérescence discale associée. Ceci survient chez un individu âgé au moins sur le plan physiologique, et s’accompagne de signes radiculaires au premier plan, car l’arc postérieur reste en continuité du corps, et l’effet « coupe-cigare » ajouté à l’hypertrophie arthrosique des articulations inter-apophysaires crée un syndrome canalaire de type claudication neurologique ou de type radiculalgique, selon la prédominance globale ou latérale de la sténose. Le déplacement lui-même est habituellement modéré.
- Le type IV intéresse le glissement acquis à la suite de la rare fracture traumatique des apophyses articulaires.
- Le type V comprend les conditions pathologiques locales (infection, tumeur) ou générales conduisant à la destruction des facettes, pédicules ou isthmes.
- Enfin le type VI, ajouté ultérieurement, est le spondylolisthésis créé par la destruction chirurgicale de l’arc postérieur.
Physiopathologie
Les facteurs locaux sont essentiels : hyper lordose constitutionnelle, répétition de mouvements en hyper-lordose, angle d’incidence élevé, apophyses articulaires inférieures de grand volume à l’avant dernier niveau mobile, mettent l’isthme aplati du dernier niveau mobile en danger de cisaillement avec les apophyses articulaires sacrées à la faveur d’un traumatisme aigu ou chronique.
Plateau sacré arrondi « en dôme », incidence élevée, sont des facteurs d’aggravation du glissement.
Des facteurs congénitaux et héréditaires inconnus ont été évoqués pour expliquer la fréquence accrue dans certaines populations et certaines familles, sans qu’on connaisse le rôle de la posture et de l’habitus.
La spondylolyse serait une pathologie de la marche, inconnue chez les patients non ambulatoires, bien que des cas aussi précoces qu’à l’âge de 2 ans aient pu être décrits.
Lorsque l’isthme est rompu ou allongé de façon bilatérale, le corps vertébral libéré du contrôle de l’arc postérieur soumet le disque sus-jacent et les ligaments ilio-lombaires à la totalité des contraintes de cisaillement qu’il subit. Des contraintes en cisaillement et de la capacité de résistance de l’anulus dépendent la constitution ou non d’un spondylolisthésis.
Spondylolyse et spondylolisthésis isthmique sont des aspects différents d’une affection dont la variabilité clinique est remarquable.
Il y a des spondylolyses bien tolérées sur de très longues périodes sans spondylolisthésis, des spondylolisthésis sans spondylolyse, des spondylolisthésis stables non évolutifs pendant de nombreuses années, des formes d’aggravation lente ou rapide qu’il faut détecter à temps, et des formes mixtes. Il n’y a donc pas de filiation stricte entre les deux états et seule la surveillance est à même d’affirmer l’évolutivité vers l’aggravation du déplacement vertébral.
Les facteurs de progression sont : le sexe féminin, l’obésité, les contraintes en hyper lordose, l’âge (adolescents en période de croissance), la cyphose locale.
Le glissement constitué, le corps vertébral subit deux types de déplacements : le glissement vers l’avant proprement dit, la bascule en cyphose. S’y ajoute le déplacement vers le bas par effet de pente, et la ptose lorsque le corps vertébral échappe en bout de course au support du plateau vertébral érodé sous-jacent, sous l’effet du poids du tronc et de la traction du psoas sur le rachis lombaire.
Quantification du glissement
L’index de Taillard mesure le glissement à partir du repère du coin postéro-inférieur de la vertèbre olisthésique, en pourcentage de la longueur antéro postérieure du plateau sacré sous-jacent. En France, on divise traditionnellement le plateau sacré en trois tiers dans le sens antéropostérieur, et on cote le glissement antéropostérieur en fonction de l’épaisseur de ce plateau :
- Grade 0 (pas de glissement, ou spondylolyse sans glissement)
- Grade I glissement inférieur à 1/3 du plateau vertébral
- Grade II de 1/3 à 2/3, bord postérieur du corps vertébral en regard du milieu du plateau sacré
- Grade III plus de 2/3
- Grade IV glissement complet au-delà de la limite antérieure du plateau sous-jacent avec spondyloptose
Dans le monde anglo-saxon, à partir de la même référence (Meyerding), on divise le plateau sacré en quatre quarts et il y a 5 stades numérotés de I à V… Cependant, on s’accorde sur le fait que les spondylolisthésis dont le glissement (index de Taillard) est inférieur à 33 % ont une évolution et des implications thérapeutiques différentes de ceux dont le glissement est plus important. Dans les spondylolisthésis de haut grade, la partie antérieure du plateau vertébral érodée prend un aspect arrondi ou oblique, avec un plateau à double pente, qui rend la mesure de l’index de Taillard peu précise. Il existe une spondyloptose lorsque le coin postéro-supérieur du corps vertébral migré se trouve en position debout plus bas que le coin postéro-supérieur du plateau vertébral sous-jacent quel que soit le déplacement angulaire.
Clinique
Il existe en pratique 3 formes cliniques bien différentes : la spondylolyse isolée, le spondylolisthésis grade I, les spondylolisthésis grade II et III. La spondyloptose pose quant à elle des problèmes très particuliers.
Spondylolyse isthmique – Clinique
Le tableau complet est fait de douleurs lombaires diurnes ou nocturnes mais plutôt mécaniques, de radiculalgies et pseudo radiculalgies de topographie S1 plutôt que L5, siégeant dans la face postérieure des cuisses et souvent tronquées au genou, de troubles de la statique avec rétraction des ischio jambiers, dont l’origine précise est inconnue. Il est rare, mais évocateur chez un adolescent ou un adulte jeune. En pratique, nombre de spondylolyses sont de découverte systématique en l’absence complète ou presque complète de symptomatologie. Le problème est plutôt de déterminer la responsabilité d’une lyse isthmique bien banale chez un adulte lombalgique chronique afin d’éviter une chirurgie qui ne résoudra pas le problème douloureux.
Cliniquement, on recherche un trajet radiculaire vrai, un Lassègue, l’existence d’un déficit et d’une rétraction des ischio jambiers que l’on cote en mesurant l’angle poplité à 90° de flexion de cuisse en position couchée.
Spondylolyse isthmique – Imagerie
Faire le diagnostic : les clichés standard peuvent montrer la lyse à un œil entraîné, les clichés centrés de 3/4 font toujours partie de l’arsenal diagnostic à la recherche de la décapitation du petit chien radiologique de Lachapelle. Les coupes de scanner peuvent aider mais peuvent aussi passer dans le plan de la lyse et être difficiles à interpréter, quelquefois ce sont seulement les reconstructions tridimensionnelles dans le plan des isthmes qui permettent de trancher. Dans certaines formes infra radiologiques en particulier chez le jeune sportif lombalgique, la scintigraphie osseuse au Tc 99 peut faire suspecter la lyse en montrant une fixation uni ou bilatérale. La scintigraphie permet également de suspecter le caractère récent d’une lyse constituée chez un patient jeune. Le diagnostic précoce a un intérêt si on peut par un traitement précoce (diminution des activités, immobilisation) éviter de passer au stade de lyse radiologiquement constituée. Chercher autre chose : chez un adulte lombalgique chronique ou qui se plaint de radiculalgies, la lyse isthmique est l’arbre qui cache la forêt … hernie discale intra ou extra canalaire, au même niveau ou à un autre niveau, tumeurs rachidiennes voire ostéome ostéoïde sont des diagnostics associés possibles, en marge de la lombalgie essentielle d’origine discale pure dont la physiopathologie et les indications thérapeutiques restent discutées. Bilan préthérapeutique : L’IRM montre le volume du “ nodule de Gill ” , une anomalie radiculaire associée, une hernie discale au même niveau (en se méfiant des effets de volume partiel qui sont beaucoup plus fréquents), l’état du disque, et celui du disque sus jacent [59] dans une perspective chirurgicale.
Spondylolyse isthmique – Formes cliniques
La spondylolyse aiguë post traumatique, peut être unilatérale, il s’agit d’une vraie fracture en hyper extension et rotation du tronc, elle justifie un traitement conservateur prolongé dont le but est l’obtention de la consolidation et la restitution ad integrum. Devant une lyse constituée dans un cadre traumatique, la scintigraphie permet d’affirmer le caractère récent et de traiter en conséquence. L’immobilisation par corset est contraignante, car elle nécessite de prendre une cuisse de façon permanente.
Les spondylolyses strictement asymptomatiques ne justifient pas d’autre traitement qu’une surveillance d’autant plus rapprochée qu’il s’agit d’un individu jeune surtout chez l’adolescent sportif. En particulier, il n’y a pas de nécessité d’interdire les activités sportives. Il faut revoir le patient tous les trois à six mois initialement afin d’évaluer la stabilité. Le facteur d’évolutivité essentiel est le jeune âge du patient, enfant ou adolescent.
Indications thérapeutiques
Les lombalgies pures sont en général accessibles au traitement médical assorti d’exercice quotidien. Le but est de maintenir et renforcer la tonicité des muscles spinaux, et de lutter contre l’apparition d’une rétraction des ischio jambiers. Le traitement chirurgical se discute en fonction de la gêne provoquée : fréquence des épisodes de lombalgie récurrente, présence de radiculalgies, rétraction des ischio jambiers.
Techniques chirurgicales
La réparation directe du défect par greffon isthmique et instrumentation spécifique sans arthrodèse s’adresse à des patients qui ne nécessitent pas d’exploration canalaire donc sans radiculalgies vraies, qui ont un disque sous-jacent en bon état vérifié par l’IRM, et un défect isthmique de petit volume accessible en extra canalaire à un greffon osseux. Avec un tel niveau d’exigence, peu de patients justifient la chirurgie, et les indications sont limitées dans la forme actuelle. L’arthrodèse postéro-latérale non instrumentée (Wiltse) est réalisée par une double voie postéro-latérale entre longissimus et multifidus, les greffons de crête iliaque sont apposés au contact des transverses et articulaires de L4 à l’aileron sacré avivés. L’immobilisation postopératoire en appareil de contention est à mettre en œuvre jusqu’à consolidation, habituellement 3 mois. L’arthrodèse L5 S1 instrumentée après libération canalaire : après exposition et résection de l’arc postérieur mobile, on excise de façon bilatérale le nodule fibreux au contact de la racine L5 et on complète la libération par la régularisation de la partie basse du pédicule L5 qui a fréquemment une forme en crochet agrippant la racine. Une instrumentation pédiculaire est mise en place de L5 à S1 et l’arthrodèse réalisée de la transverse de L5 à l’aileron sacré après un avivement qui doit être soigneux. L’arc postérieur réséqué peut fournir assez de greffons, cependant la fréquence des non-consolidations est fonction inverse du volume et de la qualité de la greffe, ce qui justifie la prise de greffe iliaque en sous aponévrotique par la même voie d’abord. Combinaisons : la voie postéro-latérale de Wiltse permet au besoin de mettre en place une instrumentation pédiculaire qui soutiendra la greffe en cas d’hyper mobilité. L’arthrodèse postéro-latérale instrumentée peut être réalisée sans exploration endocanalaire et sans résection de l’arc postérieur, en fonction des besoins d’exploration et de stabilisation. Les données qui entrent dans le choix technique sont essentiellement l’âge (consolidation plus facilement obtenue chez l’adolescent que chez l’adulte [118], qui fait préférer une arthrodèse instrumentée), la nécessité d’une exploration endocanalaire (radiculalgies vraies, conflit endocanalaire concordant) et l’état du disque sus-jacent vérifié sur l’IRM. La dégénérescence de celui-ci engage à étendre l’arthrodèse à l’avant dernier niveau mobile.
Gastro-entérite
Une gastro-entérite est une infection inflammatoire du système digestif pouvant entraîner de la nausée, des vomissements, des crampes abdominales, des flatulences et de la diarrhée, ainsi que de la déshydratation, de la fièvre et des céphalées (maux de tête).
La gastro-entérite peut être d’origine bactérienne, c’est-à-dire due à la consommation d’eau ou de nourriture contaminée par des bactéries, telles que les colibacilles présents dans les selles. Des symptômes de gastro-entérite peuvent aussi être dus à des parasites internes, protozoaires ou amibes pathogènes, tel qu’Entamoeba histolytica provoquant la dysenterie amibienne ou amibiase généralement due à des installations sanitaires absentes ou inadéquates.
Cependant, dans plus des deux tiers des cas, elle est causée par des virus comme les rotavirus, (provoquant en particulier la gastro-entérite infantile), les norovirus (dont le virus de Norwalk), les adenovirus, des calicivirus et des astrovirus. La gastro-entérite est communément appelée « grippe intestinale » (terme inadéquat mais très répandu), lorsqu’elle est causée par un virus, et « empoisonnement alimentaire » ou plus justement « intoxication alimentaire » lorsque causée par une bactérie.
La gastro-entérite peut également révéler une dysenterie des voyageurs, aussi connue sous le terme de diarrhée du voyageur ou tourista. Celle-ci peut être due à une infection, le plus souvent par des bactéries (notamment Salmonella, Aeromonas, Escherichia coli, Campylobacter, Shigella, Vibrio cholerae (causant le choléra) et Vibrio (non cholérique). Parfois, l’infection peut provenir de virus, norovirus (principalement virus de Norwalk), adénovirus, astrovirus, entérovirus, rotavirus. Plus rarement, la tourista sera causée par des amibes ou protozoaires parasitaires (Cyclospora, Entamoeba histolytica, Cryptosporidium, Giardia ou autre parasite intestinal).
La diarrhée s’accompagne souvent de vomissements et de poussées de fièvre, mais les symptômes varient en fonction des individus. En effet, certains se contentent de vomir, d’autres n’ont aucun symptôme, et certains n’ont que la diarrhée. Si elle est trop importante, elle peut mener à une déshydratation de l’organisme.
Si la diarrhée perdure, elle peut laisser des séquelles sur la paroi intestinale, menant à une pathologie appelée syndrome de malabsorption.
Épidémiologie
Le rotavirus est la cause la plus courante de diarrhée et de déshydratation chez l’enfant, en particulier dans les pays développés. Dans le monde, on estime que 125 millions de diarrhées sont provoquées annuellement par ce virus (soit plus de 1 900 cas pour 100 000 habitants). On estime que chaque année, 800 000 personnes meurent de gastro-entérite dans le monde, dont 500 000 enfants de moins de cinq ans, ce qui représente 25 % des morts par diarrhées et 6 % des morts de moins de cinq ans.
Aux États-Unis, on estime que le rotavirus touche 80 % des enfants de moins d’un an ; chaque année, 500 000 enfants doivent faire l’objet de soins médicaux, et 50 000 doivent être hospitalisés.
En France, lors du pic de l’épidémie hivernale 2005–2006, on estime que 1 850 000 personnes ont consulté leur médecin généraliste en 8 semaines pour une gastro-entérite ; l’incidence a été de 367 cas pour 100 000 habitants (le seuil épidémique étant fixé à 279 cas pour 100 000 habitants). La surveillance de l’évolution de l’incidence en France est effectuée par le réseau Sentinelles de l’Inserm qui publie ces données, comme une société de communication spécialisée qui met aussi ces données à la disposition du public.
Il s’agit donc d’un important problème de santé publique. D’autant que chaque année, l’épidémie de gastro-entérite à rotavirus concorde souvent avec les épidémies de bronchiolite et de grippe, pouvant mettre en difficulté les systèmes de soins pédiatriques.
On peut remarquer que les gastro-entérites virales sont en recrudescence pendant l’hiver, surtout en Amérique du Nord et en Europe.
Diagnostic
Les symptômes habituels de la gastro-entérite sont des nausées, la perte d’appétit, crampes abdominales et vomissements qui apparaissent brutalement, de la diarrhée, de la fièvre et des céphalées (maux de tête). Plus rarement, des vertiges et une hypotension peuvent accompagner les symptômes, sans doute liés à la déshydratation et à la fatigue.
Les symptômes communément associés à la gastro-entérite, c’est-à-dire principalement les vomissements et la diarrhée, peuvent également être signes d’un empoisonnement (fruits de mer, champignons toxiques) ou d’infections systémiques (pneumonie, septicémie, etc.). Par un interrogatoire précis et le contexte clinique, il sera possible d’éliminer ces hypothèses.
La gastro-entérite peut parfois déboucher sur des complications telles que la déshydratation, pouvant même conduire à une hospitalisation. Les personnes à risque sont les jeunes enfants et les nourrissons, les personnes âgées, et les personnes ayant un système immunitaire affaibli par une maladie (VIH par exemple). Les signes de la déshydratation sont une sécheresse de la peau et de la bouche, les yeux et les parties molles du crâne (chez les nourrissons) enfoncées, des faiblesses, crampes, et perte de poids, et des urines moins fréquentes et plus foncées que d’habitude.
Si l’on suspecte une gastro-entérite d’origine bactérienne, il est possible d’effectuer une analyse des selles au laboratoire (coproculture) à la recherche de la bactérie en cause.
Génétique
Une mutation sur le gène FUT2 (en), présente chez 20 % des Européens, confère une haute résistance, voire une immunité, contre le norovirus, responsable à 85 % des gastro-entérites non bactériennes, en dehors du jeune enfant.
Antibody List
| 1-21.2 | cAMP-specific phosphodieserase PDE4A4 |
| 10.1.1 | Chloride channel calcium activated 3A1; CLCA1 protein |
| 103BF1.1 | Tannerella forsythia, strain FDC 331 (OMZ 348) |
| 10C1 (reovirus) | reovirus outer capsid protein sigma 3 |
| 10C9 anti-late bloomer | late bloomer |
| 10D3 anti-wrapper | wrapper |
| 10E5 | integrin alphaIIb |
| 10F1 | ecdysone receptor (EcR) (common), Manduca sexta |
| 10F5 | myosin heavy chain, fast, 2B |
| 10F6 (reovirus) | reovirus capsid protein mu 1C |
| 10G10 (reovirus) | reovirus outer capsid protein sigma 3 |
| 116BF1.2 | Tannerella forsythia, strain FDC 331 (OMZ 348) |
| 12-120-94/6 | csA (contact site A glycoprotein) |
| 12/101 | skeletal muscle marker, 102 kDa |
| 12/21/1-C-6 | proteoglycan, hyaluronic acid binding region; aggrecan |
| 123-353-1 | csA (contact site A glycoprotein) |
| 12A7 | Frizzled2 |
| 12C5 | versican (hyaluronate-binding region) |
| 12F10-5F11 | Megator |
| 12G10 anti-alpha-tubulin | tubulin (alpha-) |
| 12G9 | filament antigen |
| 130-80-2 | crystal protein |
| 132-250-1 | Peptide II B 80kD |
| 135-409-16 | Cap32/34 |
| 138AA1.1 | Aggregatibacter actinomycetemcomitans serotype c |
| 13C9 anti-Robo | Robo (Drosophila) |
| 13F4 | muscle marker |
| 13G3B7 Fibronectin III-15 | fibronectin (III-15) |
| 141AA1 | Aggregatibacter actinomycetemcomitans serotype b |
| 145AA1.1 | Aggregatibacter actinomycetemcomitans serotype b |
| 14A3D2 | Mmp1 hemopexin domain |
| 14C9 anti-Robo3 extracellular | Robo3 extracellular (Drosophila) |
| 14a9 | nuclear lamins II/III (Xenopus) |
| 14h7 | vimentin (Xenopus) |
| 15.3B9(NOT1) | notochord marker |
| 150AA1.1 | Aggregatibacter actinomycetemcomitans serotype a |
| 151-IgG or 151-8 AE4 | epidermal growth factor receptor |
| 151AA1.2 | Aggregatibacter actinomycetemcomitans serotype a |
| 159-183-10 | Proteasome subunit |
| 159-291-1 | actin binding protein 34 (ABP34) |
| 15AE10 | integrin beta-3 (CD61), human |
| 15C3 | ecdysone receptor (EcR) (common), Manduca sexta |
| 15F12F8 | ribosome, 30S (E. coli) |
| 15G1a(EcR-A) | ecdysone receptor (EcR-A) |
| 15H2 anti-Robo3 cytoplasmic | Robo3 cytoplasmic (Drosophila) |
| 16-3C1 | transketolase |
| 16.5H2 | neuronal, motor, marker (SC-1) |
| 169-477-5 | talin |
| 169-90-2 | actin binding protein 34 (ABP34) |
| 16B2B1 | ribosome, 30S (E. coli) |
| 17.1G6 | cardioactive peptide B |
| 171-337 | Proteasome subunit 5 |
| 176-2-5 | coronin |
| 176-3-6 | coronin |
| 177-29-B2/5 | Coactosin p17 |
| 179ME1 | Mutanase G |
| 191ME1 | Mutanase G |
| 193E11E5B11 | ribosome, (E. coli) |
| 193F10C8C11 | ribosome, (E. coli) |
| 194-62-6 | Coronin N-terminal |
| 195ME3 | Mutanase G |
| 19C2 anti-Sema II | Sema II |
| 1A1-3C2 | Skeletor |
| 1A10 | oncomodulin (beta-parvalbumin) |
| 1A8 | Us9 (pseudorabies virus) |
| 1B1 | hu-li tai shao, not hts-PC |
| 1B11 GPI-linked Neurocan receptor | neurocan receptor |
| 1B2 JW | hexokinase (Type I isozyme) |
| 1B5 | integrin alphaIIb/beta-3 (CD61) complex, mouse |
| 1B6-C4 | blastema, regenerating |
| 1C10 | HNK-1 epitope |
| 1C11 | Frizzled protein |
| 1D1 | neurocan (C-terminal epitope) |
| 1D10 | CD44 |
| 1D12 | Gurken protein |
| 1D4 anti-Fasciclin II | fasciclin II (Drosophila) |
| 1D4B | LAMP-1 (110 kDa lysosomal membrane glycoprotein) |
| 1D8 (reovirus) | reovirus sigma NS |
| 1D9-E11 | blastema, regenerating |
| 1E11 | synaptotagmin B |
| 1E12 | actinin, smooth muscle alpha |
| 1E8 | Schwann cell marker, P(o) (avian) |
| 1F6 | neurocan (N-terminal epitope) |
| 1G12 | nidogen/entactin |
| 1G2 | FXR2 |
| 1G9 | hindsight protein |
| 1H6-E9 | blastema, regenerating |
| 1H9 Fibronectin Hep2 / 1H9B2 | fibronectin (III-14 in Hep2) |
| 1h5 | cytokeratin type II (Xenopus) |
| 20-121-1 | csA (contact site A glycoprotein) |
| 20-53-1 | csA (contact site A glycoprotein) |
| 200MH1 | Mutanase G |
| 20B4 | neural crest cells |
| 20H5 | Cdk7, recombinant |
| 21-55-4 | myosin II heavy chain |
| 21-96-3 | myosin II heavy chain |
| 210-183-1 | Fimbrin (p67) |
| 210-287-3 | Fimbrin |
| 212WR2 | Campylobacter rectus |
| 21A6 | retinal space (mechanoreceptors?) |
| 22/18 | blastemal regeneration cell marker of the newt |
| 224-236-1 | actin |
| 224-256-2 | V_H_ATpase c-subunit |
| 227-341-4 | talinA (N-terminal) |
| 22B12B2 | 30S ribosomal protein S3 |
| 22C10 | futsch |
| 22d4 | mannose 6-phosphate receptor, cation-dependent |
| 23-132-27 | calmodulin |
| 23.4-5 | neuronal marker (TAG-1) |
| 232-238-10 | Cortexillin II |
| 232-238-8 | Cortexillin II |
| 23A7 (anti NrCAM) | NrCAM |
| 23C7 anti-wit | wit |
| 23E9E7 | ribosome, 30S (E. coli) |
| 24 | ATPase, (Na(+) K(+)) beta-subunit |
| 24-210-2 | csA (contact site A glycoprotein) |
| 241-438-1 | Cortexillin I |
| 248AA4.4 | Aggregatibacter actinomycetemcomitans serotype e |
| 24B10 | chaoptin, sensory neurons |
| 252-234-2 | calreticulin, recombinant Dictyostelium |
| 260AA3 | Aggregatibacter actinomycetemcomitans serotype a´ |
| 261AA2.2 | Aggregatibacter actinomycetemcomitans |
| 262AA1.1 | Aggregatibacter actinomycetemcomitans serotype a´ |
| 266AA1 | Aggregatibacter actinomycetemcomitans serotype e |
| 26E4C7 | PTP-ER |
| 270-390-2 | Calnexin |
| 28F12 | TSLP |
| 2A1 | Cubitus interruptus |
| 2A12 | tracheal system; GASP |
| 2A4 | hexokinase (Type I isozyme) |
| 2A9 (reovirus) | reovirus sigma NS |
| 2B10 | Cut protein product |
| 2B49 | phosphacan/protein tyrosine phosphatase – z/b |
| 2B6 | hexokinase (Type I isozyme) |
| 2B8 | even-skipped protein |
| 2B9 | collagen type IX |
| 2C11-2 | dynein heavy chain |
| 2C2 | collagen type IX |
| 2C4-C2 | blastema, regenerating |
| 2C6 | alpha-2-macroglobulin receptor |
| 2D5 | MHC class II |
| 2D7F8 | PTP-ER |
| 2E8 | laminin |
| 2E9 | myosin heavy chain, neonatal fast |
| 2F4 | myosin heavy chain, fast, jaw muscle specific |
| 2F4 (reovirus) | reovirus sigma 1s |
| 2F5 (reovirus) | reovirus sigma NS |
| 2F5-1 | FMRP (residues #1-204) |
| 2F7 | myosin heavy chain, fast, 2A |
| 2G9 | LAMP (gene symbol: Lsamp) |
| 2H3 | Neurofilament (NF-M) |
| 2H7 (reovirus) | MBP-sigma NS-His |
| 2H8 | CD31 (PECAM-1) |
| 3.1C12 | neuronal marker (TAG-1) |
| 31 or 31-2 | laminin |
| 32-5B6 | S-adenosylhomocysteine hydrolase |
| 325AA2 | Aggregatibacter actinomycetemcomitans |
| 326PM2 | Parvimonas micra |
| 328AA2.2 | Aggregatibacter actinomycetemcomitans serotype d |
| 33 or 33-2 | heparan sulfate proteoglycan |
| 33-294-17 | csA (contact site A glycoprotein) |
| 330AA3.2 | Aggregatibacter actinomycetemcomitans serotype d |
| 341AN3 | Actinomyces oris |
| 34B3 anti-Fasciclin II | fasciclin II (Drosophila, all isoforms) |
| 34C | Ryanodine receptor |
| 37-1A9 | Xenopus nuclear factor, xnf7 |
| 37-1B2 | transketolase |
| 371D12F6 | ribosome, 30S (E. coli) |
| 373C9C3A1 | ribosome, 30S (E. coli) |
| 389C6G7B9 | ribosome, 30S (E. coli) |
| 39 | collagen type VI |
| 39.3F7 | Islet-1 specific homeobox |
| 39.4D5 | Islet-1 & Islet-2 homeobox |
| 390C3F1D3 | ribosome, 30S (E. coli) |
| 391B9E4E1 | ribosome, 30S (E. coli) |
| 396AN1 | Actinomyces naeslundii, oris and israelii |
| 397AN1.1 | Actinomyces naeslundii and oris |
| 39BI1.1.2 | Prevotella intermedia and nigrescens (cell surface antigen) |
| 3A10 | neurofilament-associated antigen |
| 3A6 | protein tyrosine phosphatase, receptor-linked, DPTP99A |
| 3A6B4 | Mmp1 catalytic domain |
| 3A9 (323 or M10-2) | spectrin, alpha |
| 3B11 anti-grasshopper Fas I | fasciclin I (grasshopper) |
| 3B2 | collagen type III |
| 3B3 | decorin |
| 3B5 | AP-2 alpha |
| 3B8D12 | Mmp1 catalytic domain |
| 3C10 anti-even skipped | even-skipped protein (Drosophila) |
| 3C11 (anti SYNORF1) | synapsin |
| 3CB2 | radial glial cells / vimentin |
| 3E10 (reovirus) | reovirus sigma NS |
| 3E2 (reovirus) | MBP-sigma 1s |
| 3F11 | protein tyrosine phosphatase 69D |
| 3F2/D8 | Podocalyxin/gp135 |
| 3F8 | phosphacan/protein tyrosine phosphatase – z/b |
| 3H1 | keratan sulfate (brain) |
| 3H11 | laminin |
| 3H1D8F5 anti-gigas | gigas gene product |
| 3H2 2D7 | synaptotagmin |
| 3H5 | HNK-1 epitope |
| 3gA2 | lysine decarboxylase (E. corrodens) |
| 3hE5 | lysine decarboxylase (E. corrodens) |
| 4-34.10 | cAMP-specific phosphodieserase PDE4A4 |
| 40-1a | beta-galactosidase |
| 40.2D6 | Islet-1 homeobox |
| 40.3A4 | islet-1 homeobox |
| 40BI3.2.1 | Prevotella intermedia (cell surface antigen) |
| 40E-C | radial cells and radial glial cells (vimentin) |
| 421AG1 | Actinomyces naeslundii and oris |
| 455-2A4 | Transcription factor elt-2 |
| 45E10 | protein tyrosine phosphatase, receptor-linked, DPTP10D |
| 47-18-9 | alpha-actinin |
| 47-60-8 | actinin, alpha-47-60-8 |
| 47-62-17 | alpha-actinin |
| 49BG1.3 | Porphyromonas gingivalis (cell surface antigen) |
| 4A3 (reovirus) | reovirus capsid protein mu 1C |
| 4A5 | major sperm protein (MSP) |
| 4A6 | myosin heavy chain, fast, extraocular specific |
| 4A6 JW | hexokinase (Type I isozyme) |
| 4B12 | Fibronectin |
| 4B5.13 | cAMP-specific phosphodieserase PDE4A8 |
| 4C1 | transglutaminase, tissue |
| 4C5 | rhodopsin 1 (Rh1 Drosophila) |
| 4C7 | HNF3b / FoxA2 |
| 4C71B7 | protein tyrosine phosphatase, receptor-linked, DPTP99A |
| 4C9H4 anti-peanut | peanut gene protein product |
| 4D4 | Wingless protein |
| 4D6 | collagen type IX |
| 4D7/TAG1 | neuronal marker (TAG-1) |
| 4D9 anti-engrailed/invected | engrailed/invected gene products |
| 4E1.16 | cAMP-specific phosphodieserase PDE4A8 |
| 4E2(3G2) | neuromuscular junction and reactive Schwann cell associated antigen |
| 4F12 | sialomucin complex (Muc4) |
| 4F2 | Lim 1+2 / LhxV5 |
| 4F2 (reovirus) | reovirus |
| 4F3 anti-discs large | discs large (Drosophila) |
| 4G1 | MSX1+2 |
| 4G11 | Engrail-1 |
| 4H2 | Fibronectin |
| 4H6 | neurofilament (NF-M) |
| 4d | NCAM (cytoplasmic domain) |
| 5-13.4 | cAMP-specific phosphodieserase PDE4D5 |
| 5-14 | myosin heavy chain A |
| 5-23 | paramyosin |
| 5-6 | myosin heavy chain A |
| 5.1H11 | NCAM |
| 50.5A5 | LMX |
| 51.4H9 | Islet-2 |
| 54-11-10 | Hisactophilin, Dd gelation factor |
| 56-396-5 | Myosin II |
| 56.4H7 | rhoB |
| 5A3 | hexokinase (Type I isozyme) |
| 5A5 | NCAM (sialylated form); PSA-NCAM |
| 5A6 | tau |
| 5B1-E6 | blastema, regenerating |
| 5B5-2 | Hoxc9 |
| 5B8 | NCAM |
| 5C2 | hexokinase (Type I isozyme) |
| 5C3 (reovirus) | reovirus outer capsid protein sigma 3 |
| 5C6 | collagen type VI |
| 5C6 (reovirus) | reovirus |
| 5C9 | perlecan (domain IV) |
| 5D2-27 | Pgp-1 (Ly-24) lymphocyte cell adhesion glycoprotein |
| 5D3 | cadherin, E- |
| 5E1 | sonic hedgehog |
| 5E12-E3 | blastema, regenerating |
| 5E4 | AP-2 alpha |
| 5F10 | allatostatin (Ast7) |
| 5G10 | lysosomal membrane glycoprotein (LEP-100) / CD107 |
| 5G12-10 | Hoxc10 |
| 5G2 anti-enabled | enabled |
| 5H7B11 | Mmp1 catalytic domain |
| 5e | NCAM (extracellular domain) |
| 602.29 cl. 11 | chromosome 12, 21 kDa product |
| 60BG1.3 | Porphyromonas gingivalis, strain W83 (OMZ 309) |
| 61BG1.3 | Porphyromonas gingivalis, strain W83 (OMZ 309) |
| 62.1E6 | Slug |
| 65B12 | TSLP |
| 67.4E12 | Lim 3 |
| 6B3 | Cadherin, N- (neural) |
| 6B6 | cadherin, C- |
| 6B7 | ecdysone receptor (EcR) B1-isoform specific, Manduca sexta |
| 6B9 | Villin-like protein quail; 6B9 |
| 6C4 | collagen type XVIII |
| 6C8-A2 | blastema, regenerating |
| 6D1 | GPIb; von Willebrand factor receptor; GPIb/IX complex |
| 6D2 | agrin |
| 6D6 | decorin |
| 6E8 | Posterior sex combs protein |
| 6F1 | integrin alpha-2; alpha2beta1 integrin complex |
| 6F8 anti-grasshopper Sema I | Sema I (grasshopper) |
| 6G10-2C7 | blastema, regenerating |
| 6G7 | tubulin |
| 6H1 | myosin heavy chain, fast, 2X |
| 6H4 anti-highwire | highwire |
| 70-100-1 | porin |
| 72.5B10 | ER81 |
| 74.5A5 | Nkx2.2 |
| 7A1 (reovirus) | reovirus outer capsid protein sigma 3 |
| 7A2 GPI-linked Neurocan receptor | neurocan receptor |
| 7A6 | NFATc1 |
| 7B1 | decorin |
| 7B11 | cyclic nucleotide-gated channel (rOCN2, rat) |
| 7D6 | L-CAM, E-cadherin, uvomorulin |
| 7E2 | integrin beta-1 |
| 7E9 | fibrinogen |
| 7F4 (reovirus) | reovirus gamma 2 vertex protein |
| 7G1-1 | FMRP |
| 7G10 anti-Fasciclin III | fasciclin III (Drosophila) |
| 7G4 | visinin |
| 7H2 | integrin beta-3 |
| 8.1.1 | stromal cell line (murine) |
| 80-52-13 | Discoidin I (cAMP binding domain) |
| 81.5C10 | MNR2/HB9/Mnx1 |
| 86f7 | mannose 6-phosphate / IGF II receptor, cation-independent |
| 8A2 | neural associated ganglioside |
| 8B22F5 | protein tyrosine phosphatase, receptor-linked, DPTP10D |
| 8B4D2 (MH2B) | glutamate receptor subunit, DGluR-IIA |
| 8C2 | cadherin, E- |
| 8C3 | syntaxin (Drosophila) |
| 8C6 anti-grasshopper Fas II | fasciclin II (grasshopper) |
| 8C8 | integrin beta-1 (Xenopus) |
| 8D12 anti-Repo | Repo |
| 8D9 | L1-like CAM (avian) |
| 8F12 (reovirus) | reovirus outer capsid protein sigma 3 |
| 8F3 | chicken cell marker |
| 8H1 (reovirus) | reovirus sigma NS |
| 8H6 (reovirus) | reovirus capsid protein mu 1C |
| 8H9 | CD64 |
| 8e6 | talin (avian) |
| 9 D10 | titin |
| 9.1 ITGA7 | Integrin alpha-7, extracellular domain |
| 9.4A anti-Trio | Trio |
| 9/30/8-A-4 | link protein |
| 99.1-3A2 | Evx1 |
| 9B2.1 anti-glass | glass Drosophila protein |
| 9B8 anti-sidestep | sidestep |
| 9B9 | ecdysone receptor (EcR) (common), Manduca sexta |
| 9BA12 | chondroitin sulfate proteoglycans (carbohydrate epitope) |
| 9D82B3 | DLAR |
| 9E 10 | c-myc |
| 9H5 | integrin beta 3 |
| 9H6 | entactin (synaptic) |
| 9S4 | Selenoprotein P |
| A12 | cyclin A |
| A21F7 | integrin alpha-5 (avian) |
| A2B11 | transitin |
| A4.1025 | myosin (human all fibers) |
| A4.1519 | Myosin heavy chain (human nascent secondary and all fast fibers) |
| A4.74 | Myosin heavy chain (human fast fibers) |
| A4.840 | Myosin heavy chain (human slow fibers) |
| A4.951 | Myosin heavy chain (human slow fibers) |
| A6 BCM | A6 antigen in biliary epithelial/oval cells |
| A72-24 | phosphatidylinositol-specific phospholipase C |
| AA12.1 | tubulin (beta-) |
| AA4.3 | tubulin (alpha-) |
| AB8 | myosin heavy chain, adult |
| ABL-93 | LAMP-2 (110 kDa lysosomal membrane glycoprotein) |
| ABP2 | ABP2 olfactory binding protein |
| AC4 | neural tube, dorsal |
| AD4.4(EcR-B1) | ecdysone receptor (EcR-B1) |
| ADL101 | lamin Dm0 |
| ADL195 | lamin Dm0 |
| ADL40 | lamin Dm0 |
| ADL46 | lamin Dm0 |
| ADL67.10 | lamin Dm0 |
| ADL84.12 | lamin Dm0 |
| AE-2 | acetylcholinesterase |
| AF-CA2 | Aspergillus flavus isolate 93803 |
| AFFN-AKT3-10E3 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-10F2 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-13B4 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-16B3 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-18F11 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-1B9 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-1H11 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-20A3 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-2F10 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-7H5 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-8F3 | RAC-gamma serine/threonine-protein kinase |
| AFFN-AKT3-9F4 | RAC-gamma serine/threonine-protein kinase |
| AFFN-APLF-15F11 | Aprataxin and PNKP like factor |
| AFFN-APLF-4H5 | Aprataxin and PNKP like factor |
| AFFN-ARHGC-1-15A6 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-ARHGC-1-2B12 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-ARHGC-1-9H2 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-ARHGEF12-15D11 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-ARHGEF12-17B8 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-ARHGEF12-17E4 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-ARHGEF2-11E6 | Rho guanine nucleotide exchange factor (GEF) 12 |
| AFFN-BIRC3-10E9 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-10F3 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-10G6 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-10H11 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-11G3 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-11H11 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-12D12 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-12F9 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-15B3 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-16H8 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-17G5 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-19C4 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-1B11 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-2D7 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC3-9D1 | Baculoviral IAP repeat containing 3 |
| AFFN-BIRC7-11F1 | Baculoviral IAP repeat containing 7 |
| AFFN-BIRC7-11H5 | Baculoviral IAP repeat containing 7 |
| AFFN-BIRC7-2B3 | Baculoviral IAP repeat containing 7 |
| AFFN-BIRC7-3B3 | Baculoviral IAP repeat containing 7 |
| AFFN-BLK-10F9 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BLK-11H2 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BLK-14B8 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BLK-19H2 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BLK-20G3 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BLK-6A5 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BLK-9E2 | B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) |
| AFFN-BMX-10D11 | BMX non-receptor tyrosine kinase |
| AFFN-BMX-11C9 | BMX non-receptor tyrosine kinase |
| AFFN-BMX-11D11 | BMX non-receptor tyrosine kinase |
| AFFN-BMX-15D7 | BMX non-receptor tyrosine kinase |
| AFFN-BMX-18E3 | BMX non-receptor tyrosine kinase |
| AFFN-BMX-8H10 | BMX non-receptor tyrosine kinase |
| AFFN-BRCA1-14C4 | Breast cancer 1, early onset |
| AFFN-BRCA1-1G5 | Breast cancer type 1 susceptibility protein |
| AFFN-BRDT-12B2 | Bromodomain, testis-specific |
| AFFN-BRDT-13E3 | Bromodomain, testis-specific |
| AFFN-BRDT-19H3 | Bromodomain, testis-specific |
| AFFN-BTK-8A6 | Bruton agammaglobulinemia tyrosine kinase |
| AFFN-BTK-9A11 | Bruton agammaglobulinemia tyrosine kinase |
| AFFN-BTK-9F2 | Bruton agammaglobulinemia tyrosine kinase |
| AFFN-CAMKK2-10H2 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-1H3 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-2B4 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-3B11 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-3B4 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4E6 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8-B6 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8-E4 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8-G7 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8-G8 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8-H11 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-4G8-H8 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-5D6 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-7E1 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-7E7 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-7E7-D11 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-7E7-E12 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CAMKK2-8B4 | calcium/calmodulin-dependent protein kinase kinase 2, beta |
| AFFN-CBL-1G11 | Cbl proto-oncogene, E3 ubiquitin protein ligase |
| AFFN-CDC34-10D1 | Cell division cycle 34 |
| AFFN-CDC34-1G12 | Cell division cycle 34 |
| AFFN-CEP170-20B9 | Centrosomal protein 170kDa |
| AFFN-CEP170-7A12 | Centrosomal protein 170kDa |
| AFFN-CHEK2-17D2 | Serine/threonine-protein kinase Chk2; Checkpoint kinase 2 |
| AFFN-CIT-1F12 | Citron Rho-interacting kinase |
| AFFN-CIT-1G6 | Citron Rho-interacting kinase |
| AFFN-CIT-2A11 | Citron Rho-interacting kinase |
| AFFN-CIT-2D10 | Citron Rho-interacting kinase |
| AFFN-CIT-3H6 | Citron Rho-interacting kinase |
| AFFN-CSNK1E-13D7 | Casein kinase I isoform epsilon |
| AFFN-CSNK2A1-5F1 | Casein kinase II subunit alpha |
| AFFN-CSNK2A2-5F2 | Casein kinase II subunit alpha prime |
| AFFN-DLG1-4D6 | Discs, large homolog 1 (drosophila) |
| AFFN-DLG2-13F7 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-15D2 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-17C6 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-17F1 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-1B9 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-1H12 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-3A3 | Discs, large homolog 2 (drosophila) |
| AFFN-DLG2-4C3 | Discs, large homolog 2 (drosophila) |
| AFFN-DNTT-13A10 | DNA nucleotidylexotransferase |
| AFFN-DNTT-19B6 | DNA nucleotidylexotransferase |
| AFFN-DNTT-3D7 | DNA nucleotidylexotransferase |
| AFFN-DUSP7-14G9 | Dual specificity phosphatase 7 |
| AFFN-ECHS1-5F7 | Enoyl CoA hydratase, short chain, 1, mitochondrial |
| AFFN-ECHS1-7E6 | Enoyl CoA hydratase, short chain, 1, mitochondrial |
| AFFN-ECHS1-7G6 | Enoyl CoA hydratase, short chain, 1, mitochondrial |
| AFFN-ECHS1-9B3 | Enoyl CoA hydratase, short chain, 1, mitochondrial |
| AFFN-ECHS1-9E6 | Enoyl CoA hydratase, short chain, 1, mitochondrial |
| AFFN-ECHS1-9E9 | Enoyl CoA hydratase, short chain, 1, mitochondrial |
| AFFN-EEF2K-10E8 | Eukaryotic elongation factor-2 kinase |
| AFFN-EEF2K-13D2 | Eukaryotic elongation factor-2 kinase |
| AFFN-EEF2K-3E10 | Eukaryotic elongation factor-2 kinase |
| AFFN-EEF2K-3F6 | Eukaryotic elongation factor-2 kinase |
| AFFN-FASN-10H6 | Fatty acid synthase |
| AFFN-FASN-18A8 | Fatty acid synthase |
| AFFN-FASN-19C3 | Fatty acid synthase |
| AFFN-FASN-3E7 | Fatty acid synthase |
| AFFN-FASN-4A1 | Fatty acid synthase |
| AFFN-FASN-4H3 | Fatty acid synthase |
| AFFN-FASN-5H12 | Fatty acid synthase |
| AFFN-FASN-5H9 | Fatty acid synthase |
| AFFN-FASN-6E5 | Fatty acid synthase |
| AFFN-FASN-7A8 | Fatty acid synthase |
| AFFN-FASN-9H3 | Fatty acid synthase |
| AFFN-FER-18A7 | Fer tyrosine kinase |
| AFFN-FER-2G12 | Fer tyrosine kinase |
| AFFN-FER-3D4 | Fer tyrosine kinase |
| AFFN-FER-5F12 | Fer tyrosine kinase |
| AFFN-FER-7F12 | Fer tyrosine kinase |
| AFFN-FES-2F3 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-3B12 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-3D1 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-4D12 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-4G1 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-7B3 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-7C2 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-7G6 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-8D1 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-8D1-B2 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-8D1-B6 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FES-9D2 | Tyrosine-protein kinase Fes/Feline sarcoma oncogene |
| AFFN-FGR-14A10 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-14A3 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-15A5 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-19G5 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-1A7 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-1D12 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-1G12 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-4E7 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-7A2 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FGR-7G3 | Feline Gardner-Rasheed sarcoma viral oncogene homolog |
| AFFN-FH-1A7 | Fumarate hydratase, mitochondrial |
| AFFN-FH-1F6 | Fumarate hydratase, mitochondrial |
| AFFN-FH-2G8 | Fumarate hydratase, mitochondrial |
| AFFN-FUS-11E11 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-13B6 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-15H5 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-16F1 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-18B9 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-19B2 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-5A9 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FUS-6F2 | Fused in sarcomas/RNA-binding protein FUS |
| AFFN-FYN-13B9 | Tyrosine-protein kinase Fyn/ FYN oncogene related to SRC, FGR, YES |
| AFFN-FYN-18G7 | Tyrosine-protein kinase Fyn/ FYN oncogene related to SRC, FGR, YES |
| AFFN-FYN-7H11 | Tyrosine-protein kinase Fyn/ FYN oncogene related to SRC, FGR, YES |
| AFFN-GNG2-11F6 | Guanine nucleotide binding protein (G protein), gamma 2 |
| AFFN-GNG2-20G6 | Guanine nucleotide binding protein (G protein), gamma 2 |
| AFFN-GNG2-2B12 | Guanine nucleotide binding protein (G protein), gamma 2 |
| AFFN-GNG2-3B2 | Guanine nucleotide binding protein (G protein), gamma 2 |
| AFFN-GOT2-12F6 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GOT2-15C9 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GOT2-1A8 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GOT2-2F2 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GOT2-5G7 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GOT2-6B2 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GOT2-7H1 | Glutamic-oxaloacetic transaminase 2, mitochondrial |
| AFFN-GRB7-1C7 | Growth factor receptor-bound protein 7 |
| AFFN-GRB7-1H4 | Growth factor receptor-bound protein 7 |
| AFFN-GRIP1-18D11 | Glutamate receptor interacting protein 1 |
| AFFN-GRIP1-20E9 | Glutamate receptor interacting protein 1 |
| AFFN-GRIP1-9G1 | Glutamate receptor interacting protein 1 |
| AFFN-GRK5-10A12 | G protein-coupled receptor kinase 5 |
| AFFN-GRK5-17D4 | G protein-coupled receptor kinase 5 |
| AFFN-GRK5-18F6 | G protein-coupled receptor kinase 5 |
| AFFN-GRK5-2D8 | G protein-coupled receptor kinase 5 |
| AFFN-GRK5-3F12 | G protein-coupled receptor kinase 5 |
| AFFN-HSD17B10-1B4 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| AFFN-HSD17B10-1C2 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| AFFN-HSD17B10-1C9 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| AFFN-HSD17B10-1H2 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| AFFN-HSD17B10-3C9 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| AFFN-HSD17B10-4D9 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| AFFN-HSH2D-6G7 | Hematopoietic SH2 domain containing |
| AFFN-HSH2D-7B7 | Hematopoietic SH2 domain containing |
| AFFN-HSH2D-8D9 | Hematopoietic SH2 domain containing |
| AFFN-HSH2D-8E8 | Hematopoietic SH2 domain containing |
| AFFN-HSH2D-8F2 | Hematopoietic SH2 domain containing |
| AFFN-HSH2D-9E3 | Hematopoietic SH2 domain containing |
| AFFN-HSPB8-13B6 | Heat shock 22kDa protein 8 |
| AFFN-HSPB8-16F8 | Heat shock 22kDa protein 8 |
| AFFN-HSPB8-2E10 | Heat shock 22kDa protein 8 |
| AFFN-HSPB8-4D5 | Heat shock 22kDa protein 8 |
| AFFN-HSPB8-7C5 | Heat shock 22kDa protein 8 |
| AFFN-IGFBP2-10B4 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-1B4 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-1H12 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-5D1 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-5F5 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-6D11 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-7G8 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-8G8 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-9A6 | Insulin-like growth factor binding protein 2 |
| AFFN-IGFBP2-9D9 | Insulin-like growth factor binding protein 2 |
| AFFN-ILK-7C7 | Integrin-linked protein kinase |
| AFFN-INADL-1-15D1 | InaD-like protein |
| AFFN-INADL-1-3G6 | InaD-like protein |
| AFFN-INADL-1-4A11 | InaD-like protein |
| AFFN-INADL-1-5E7 | InaD-like protein |
| AFFN-INADL-1-5H2 | InaD-like protein |
| AFFN-ITK-12A9 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-12B1 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-12F4 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-14G9 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-14H5 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-16C12 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-18H8 | IL2-inducible T-cell tyrosine-protein kinase |
| AFFN-ITK-3A8 | IL2-inducible T-cell tyrosine-protein kinase |
Antigen List
| 14-3-3 sigma 26S proteasome non-ATPase regulatory subunit 4 2F7 30S ribosomal protein S3 5-bromo-2′-deoxyuridine BrdU 6XHis peptide, linear 8-oxoguanine DNA glycosylase A6 Antigen in Biliary Epithelial & Oval cells Abdominal-B homeobox protein ABP2 olfactory binding protein Abrupt acetylcholine nicotinic receptors, muscle acetylcholinesterase Achaete protein acinar, exocrine gland Acj6 actin actin associated antigen, migration-related actin binding protein 34 (ABP34) actinin, alpha- actinin, smooth muscle alpha Actinomyces naeslundii and oris Actinomyces naeslundii, oris and israelii Actinomyces oris Activating transcription factor 7; ATF7 addressin (PNAd) adducin-related protein adenovirus type 5 hexon (human) ADP-ribosylation factor-like protein 8 Aggregatibacter actinomycetemcomitans Aggregatibacter actinomycetemcomitans serotype a Aggregatibacter actinomycetemcomitans serotype a‰ Aggregatibacter actinomycetemcomitans serotype b Aggregatibacter actinomycetemcomitans serotype c Aggregatibacter actinomycetemcomitans serotype d Aggregatibacter actinomycetemcomitans serotype e Aggregatibacter actinomycetemcomitansserotype d agrin Aldo-keto Reductase Family 1 Member B1 Aldo-keto Reductase Family 1 Member C1 Aldo-keto reductase family 1 member C2 alkaline phosphatase isoenzyme alkaline phosphatase, bone and liver allatostatin (Ast7) alpha sarcoglycan alpha2bBeta3 alpha-2-macroglobulin receptor alpha-L-fucosidase aminopeptidase N ANAPC2; Anaphase promoting complex subunit 2 Anaphase promoting complex subunit 2 angiotensin converting enzyme Ankyrin repeat and BTB (POZ) domain containing 1 Ankyrin repeat and KH domain containing 1 Ankyrin repeat and zinc finger domain containing 1 Annexin A1 (Annexin I) Annexin A1 Peptide 1 Annexin A4 Peptide 1 annexin I annexins I and II Antennapedia protein Anti-silencing function 1A histone chaperone AP-2 alpha AP-3 (delta subunit) APA-2 amino acids 618-925 APEX Nuclease I apoptotic marker in phagocytic cells (nematode) Aprataxin and PNKP like factor argos-gene product ARID3A; AT rich interactive domain 3A (BRIGHT-like) Armadillo Drosophila protein Aryl hydrocarbon receptor nuclear translocator-like protein 1 Aspergillus flavus isolate 93803 ASPL-TFE3 type 1 ASPL-TFE3 type 2 AT rich interactive domain 2 (ARID, RFX-like) AT-hook-containing transcription factor Atox1 ATPase family AAA domaining containg 2 ATPase family, AAA domain containing 2 ATPase family, AAA domain containing 2; ATAD2 ATPase, (Na (+) K(+)) alpha subunit ATPase, (Na(+) K(+)) alpha-1 subunit ATPase, (Na(+) K(+)) alpha-1 subunit (avian) ATPase, (Na(+) K(+)) beta-1 subunit (avian) ATPase, (Na(+) K(+)) beta-subunit ATPase, Ca(+2) fast twitch SR (avian) / SERCA1 ATPase, Ca(+2) fast twitch SR / SERCA1 ATPase, Ca(+2) slow twitch/cardiac SR (avian), SERCA2 ATRX; Alpha thalassemia/mental retardation syndrome X-linked autoimmune double stranded DNA autoimmune single stranded DNA avian myoblastosis virus, p19^(gag) AVP axonal filaments, 56 & 58 kDa (amphibian) axons, CNS (Drosophila) B lymphoid tyrosine kinase (Tyrosine-protein kinase Blk) BACH1 BACH2; BTB and CNC homology 1, basic leucine zipper transcription factor 2 Baculoviral IAP repeat containing 3 Baculoviral IAP repeat containing 7 basement membrane marker Basic helix-loop-helix family, member a9 Basic leucine zipper transcription factor ATF-like 3 Basic leucine zipper transcription factor, ATF-like 2 Basic leucine zipper transcription factor, ATF-like 3 BATF; Basic leucine zipper transcription factor, ATF-like B-cell CLL/lymphoma 6 B-cell CLL/lymphoma 6; B-cell lymphoma 6 protein B-cell lymphoma/leukemia 11A BCL2 like 1 BCL2-like 2 BEAF Beta Catenin like protein 1 beta-defensin 3 beta-galactosidase BHK cells overexpressing mouse Glucagon-like peptide 1 receptor Bicaudal-D blastema, regenerating blastemal regeneration cell marker of the newt BMI1 polycomb ring finger oncogene; Polycomb complex protein BMI-1 BMX non-receptor tyrosine kinase Bobby sox homolog bodywall muscle cells BolA family member 3 BolA-like protein 2 bone sialoprotein II Botrytis cinerea brain-derived neurotrophic factor, fish & human brain-derived neurotrophic factor, mouse BRCA1 Interacting Protein C-Terminal Helicase 1 Peptide 1 BRD1; Bromodomain containing 1 Breast cancer 1, early onset Breast Cancer 1, Early Onset Peptide 1 Breast cancer type 1 susceptibility protein Broad (core) Broad (Z1) Broad (Z3) Bromodomain and PHD finger containing, 1 Bromodomain and WD repeat domain containing 1 Bromodomain containing 1 Bromodomain containing 3 Bromodomain containing 9 Bromodomain, testis-specific Bruchpilot Bruton agammaglobulinemia tyrosine kinase BSX; Brain-specific homeobox BTB and CNC homology 1, basic leucine zipper transcription factor 2 BTN1A1 BUB3-interacting and GLEBS motif-containing protein ZNF207 cactus cadherin, B- cadherin, C- cadherin, DE- cadherin, DN- Cadherin, E- cadherin, E- (canine) cadherin, N- cadherin, R- cadherin-6B cadherin-7 cadherin-8 calbindin-32, drosophila Calcineurin B homologous protein 2 (Hepatocellular carcinoma-associated antigen 520) calcium/calmodulin-dependent protein kinase kinase 2, beta Calcyclin (Prolactin Receptor Associated Protein) calmodulin Calnexin Calreticulin Peptide 1 calreticulin, recombinant Dictyostelium CAMP responsive element binding protein 1 cAMP responsive element binding protein 5 CAMP responsive element binding protein-like 2 cAMP-specific phosphodieserase PDE4A4 cAMP-specific phosphodieserase PDE4A8 cAMP-specific phosphodieserase PDE4D5 Campylobacter rectus Cap32/34 capping protein alpha-1 & alpha-2 subunits capping protein beta-1 subunit capping protein beta-2 subunit carbohydrate epitope, probably globoseries Carbonic anhydrase VIII cardioactive peptide B CARO2 Casein kinase I isoform epsilon Casein kinase II subunit alpha Casein kinase II subunit alpha prime CASP8AP2; CASP8-associated protein 2 Caspase 8 associated protein 2 catenin, alpha- (Drosophila) catenin, alpha N catenin, beta- catenin, p120 Caudal type homeobox 2 caveolin CBFB; Core-binding factor subunit beta Cbl proto-oncogene, E3 ubiquitin protein ligase Cbl-L protein exon 6 (aa 449-878 of L isoform) CBX2; Chromobox protein homolog 2 CCAAT/enhancer binding protein (C/EBP), beta CD1 CD11a (LFA-1 alpha subunit) CD11a (LFA-1 alpha subunit) (human) CD11a (LFA-1 alpha subunit), murine CD11b (Mac-1 alpha subunit), murine CD11b (Mac-1), murine CD11b (Mac-1, CR3) (human) CD133/ Prominin-1 CD18 (beta subunit of CD11, a, b, c), murine CD18 (beta subunit of CD11a, b, c) CD2 (LFA-2) CD21 Cd248 CD31 (PECAM-1) CD44 CD44 (CRIII) CD44 human, hyaluronate receptor CD44 molecule (Indian blood group) Peptide 1 CD45 (lymphocyte common antigen) (human) CD48 CD58 (LFA-3) CD63 (human) LAMP CD64 CD9 CDC5L; Cell division cycle 5-like Cdk7, recombinant CEBPE; CCAAT/enhancer binding protein (C/EBP), epsilon CEBPZ; CCAAT/enhancer binding protein (C/EBP), zeta Cell cycle and apoptosis regulator 2 Cell division cycle 34 Cell division cycle 34 homolog (S. Cerevisiae) cell-junction marker Centrosomal protein 170kDa chaoptin Checkpoint kinase 2 chemokine (C-X-C motif) ligand 9 chicken cell marker Chloride channel calcium activated 3A1; CLCA1 protein Chloride Intracellular Channel 1 choline acetyltransferase Choline Kinase Alpha chondroitin sulfate proteoglycans (carbohydrate epitope) Chorionic Gonadotropin, beta polypeptide Peptide 1 Chromobox homolog 1 Chromobox homolog 3 Chromobox homolog 5 Chromogranin A chromosome 1, 55-73 kDa polypeptides chromosome 11, 40 and 80 kDa polypeptides chromosome 12, 21 kDa product chromosome 19, OKa blood antigen / CD147 Citron Rho-interacting kinase Cleavage stimulation factor subunit 2 Cleavage stimulation factor subunit 2 tau variant Clec4f c-MYC CNS, leech CNS-specific antigen Coactosin p17 Coiled-coil and C2 domain containing 1A collagen (pro-) type I (aminopropeptide) collagen type II collagen type III collagen type IV collagen type IX collagen type VI collagen type X collagen type XII collagen type XVIII collagen(pro-) type I collagenase Connectin C1.427 connexin 32 COPS3; COP9 signalosome subunit 3 Coracle Core-binding factor subunit beta Core-binding factor, beta subunit coronin Coronin N-terminal Cortexillin I Cortexillin II Costal-2 (Cos2) creatine kinase Creatine kinase B chain CREB3L4; CAMP responsive element binding protein 3-like 4 CrebA transcription factor CREBL2; cAMP responsive element binding protein-like 2 Crumbs Drosophila protein crystal protein Crystallin Alpha B csA (contact site A glycoprotein) Csp CSP2 chemosensory protein CTBP1; C-terminal binding protein 1 C-terminal binding protein 1 C-terminal binding protein 2 Cubitus interruptus (Ci) full length CUGBP, Elav-like family member 2 CUGBP, Elav-like family member 4 Cut homeobox Cutoff protein, CG13190, full length protein CXXC1; CXXC finger protein 1 Cyclic AMP-dependent transcription factor ATF-2 cyclic nucleotide-gated channel (rOCN2, rat) Cyclin A cyclin B Cyclin D binding myb-like Transcription Factors 1 cyp-33E1, recombinant Cystatin A Cystatin B cysteine string protein (CSP), Drosophila cytokeratin type II (Xenopus) D4, zinc and double PHD fingers family 1 Dacapo Dachshund protein, Drosophila Dally-like protein (Dlp) DAO5 DAXX; Death-domain associated protein DAXX; death-domain associated protein DBX2; Developing brain homeobox 2 DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 Death-domain associated protein Decapping mRNA 2 decorin Deformed epidermal autoregulatory factor 1 homolog Del-1 Deleted in liver cancer 1 Delta, extracellular domain desmin dFMR1 dFMRP DGCR8 microprocessor complex subunit DHPR alpha-subunit, skeletal muscle DHPR beta subunit, skeletal muscle Dip2 dipeptidylpeptidase IV Disabled protein Discoidin I (cAMP binding domain) discs large (C. elegans) Discs large (Drosophila) Discs, large homolog 1 (drosophila) Discs, large homolog 2 (drosophila) Disp2 peptide (aa731-750 coupled to KLH) DLAR DLX3; Distal-less homeobox 3 DMPK DNA (cytosine-5-)-methyltransferase 3 alpha DNA (cytosine-5-)-methyltransferase 3-like DNA ligase 3 DNA nucleotidylexotransferase DNA repair protein XRCC1 DNA-binding protein Ikaros DNA-binding protein inhibitor ID-1 DNA-binding protein inhibitor ID-3 DNA-binding protein RFX2 DNA-binding protein RFX5 DNA-binding protein RFX6 DNA-damage inducible transcript 3 DNA-damage-inducible transcript 3 Dorsal root ganglia homeobox Dorsal, embryonic polarity protein Doublesex and mab-3 related transcription factor 2 Doublesex- and mab-3-related transcription factor B1 Doublesex protein DNA binding domain (DBD) DR1; down-regulator of transcription 1, TBP-binding (negative cofactor 2) DR1-associated corepressor DR1-associated protein 1 (negative cofactor 2 alpha) Draper Dual specificity phosphatase 7 duct, exocrine gland DVL2; Dishevelled segment polarity protein 2 DVL3; Dishevelled segment polarity protein 3 DYN-1 Dynein heavy chain Dystroglycan, alpha dystroglycan, alpha- dystroglycan, beta- dystrophin dystrophin aa 71-74 dystrophin amino acids 1749-2248 dystrophin amino acids 816-1749 dystrophin amino-acids 1749-2248 dystrophin amino-acids 1749-2248 as TrpE fusion protein dystrophin amino-acids 816-1749 dystrophin fragment encoded by exons 4-16 dystrophin fragment exons 45-50 aa 2145-2439 dystrophin fragment exons 70-79 E2F transcription factor 8 E3 ubiquitin-protein liase RNF8 E3 ubiquitin-protein ligase NEDD4-like E3 ubiquitin-protein ligase RING2 E3 ubiquitin-protein ligase SMURF1 E4F transcription factor 1 Early B-cell factor 1 ecdysone receptor (EcR common) ecdysone receptor (EcR) (common), Manduca sexta ecdysone receptor (EcR) B1-isoform specific, Manduca sexta ecdysone receptor (EcR-A) ecdysone receptor (EcR-B1) EDF1; Endothelial differentiation-related factor 1 EEA1; Early endosome antigen 1 Eef1b2 Efemp2 egg shell marker for C. elegans embryos EHMT2; Euchromatic histone-lysine N-methyltransferase 2 elav Drosophila protein elav Drosophila protein; embryonic lethal abnormal vision ELF2; E74-like factor 2 (ets domain transcription factor) ELF3; E74-like factor 3 (ets domain transcription factor, epithelial-specific ) ELK1, member of ETS oncogene family Embryonal carcinoma cell surface carbohydrate Emerin (amino acids 11-17) Emerin (amino acids 112-115 and 150-158) Emerin (amino acids 112-115) Emerin (amino acids 152-159) Emerin (amino acids 221-228) Emerin (amino acids 69-77) Emerin (amino acids 7-15) Emerin (amino acids 89-96) Enabled endoglin endoglin (CD105) endoplasmic reticulum, rough, glycoprotein Engrail-1 Engrailed/invected gene products Enolase superfamily member 1, Mitochondrial Enoyl CoA hydratase, short chain, 1, mitochondrial ENS-1; Embryonic Normal Stem Cell entactin (synaptic) EP300 EpCAM (murine) EPH receptor B4 EphB1, EphB2, EphB3 ephrin-B1 epidermal growth factor receptor epithelial stem cell marker epithelial surface marker, apical EPS15, Drosophila ER81 ERM-1, amino acids 209-563 ESRRA; Estrogen-related receptor alpha ESRRG; Estrogen-related receptor gamma estrogen receptor alpha, ligand binding domain (aa 304-554) Estrogen Receptor Binding Site Associated, Antigen 9 Estrogen-related receptor beta; Steroid hormone receptor ERR2 ETC-related transcription factor ELF-1 Ets variant 6 Ets variant 7 ETV7; Ets variant 7 Eukaryotic elongation factor-2 kinase Eukaryotic translation initiation factor 2, subunit 1 alpha Eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa Eukaryotic translation initiation factor 4A3 Eukaryotic translation initiation factor 4E Even-skipped protein Even-skipped protein (Drosophila) Evx1 Exosome complex exonuclease RRP44 Extra Sex Combs (ESC) Extradenticle protein (EXDHDcc) Eyeless protein, linker region Eyes Absent (Eya) protein Ezrin (p81) Fas (TNFRSF6) associated factor 1 fasciclin I (Drosophila) fasciclin I (grasshopper) fasciclin II (Drosophila) fasciclin II (Drosophila, all isoforms) fasciclin II (grasshopper) fasciclin II (Manduca sexta) Fasciclin III (Drosophila) Fascin Fatty acid synthase Fatty acid-binding protein, epidermal Fatty acid-binding protein, intestinal FBJ murine osteosarcoma viral oncogene homolog B Fbln2 Feline Gardner-Rasheed sarcoma viral oncogene homolog female lethal (2)D Fer tyrosine kinase FEZ family zinc finger 2 fibrillin 2-like fibrinogen fibronectin fibronectin (III-14 in Hep2) fibronectin (III-15) fibronectin, cell binding domain filament antigen Fimbrin Fimbrin (p67) Flamingo FLI1; Fli-1 proto-oncogene, ETS transcription factor floor plate marker fluorescein isothiocyanate-conjugated to keyhold limpet hemocyanin Fly Bag-of-marbles FMR1 FMRP Forkhead box A3; Hepatocyte nuclear factor 3-gamma Forkhead box B2 Forkhead box D3 Forkhead box I1 Forkhead box J2 Forkhead box N1 Forkhead box P4 Forkhead box protein P3 Forkhead box protein P4 FosB; FBJ murine osteosarcoma viral oncogene homolog B FOS-like antigen 2 Fos-related antigen 1 FOXI1; Forkhead box I1 FOXL1; Forkhead box L1 FOXM1; Forkhead box M1 FOXO3; Forkhead box O3 FOXR1; Forkhead box R1 Frizzled protein Frizzled2 Fstl3 Fukutin Fumarate hydratase, mitochondrial Fused (Fu) Fused in sarcomas/RNA-binding protein FUS futsch FXR2 G protein-coupled receptor kinase 5 GA binding protein transcription factor, beta subunit 1 GA-binding protein alpha chain ganglioside GD1a ganglioside GD1b ganglioside GM1 Ganglioside GT1b GASP (Gene Analogous to Small Peritrophins) GATA4; GATA binding protein 4 GBX-1 homeobox protein GBX1; Gastrulation brain homeobox 1 GD2 glycolipid Gelsolin gemin4 gemin5 gemin6 Geminin General transcription factor IIF subunit 2 General transcription factor IIH subunit 2 General transcription factor II-I repeat domain-containing protein 2A General transcription factors IIi germ cells, male, rodent germ-line-specific P granules gigas gene product Glass bottom boat; Protein 60A glass Drosophila protein GLI family zinc finger 1 Glial cells missing homolog 2 glial precursor Glorund Glucagon-like peptide 1 receptor Glucocorticoid modulatory element-binding protein 2 Glucose phosphate isomerase Glutamate receptor interacting protein 1 Glutamate receptor subunit, DGluR-IIA Glutamate-Cysteine Ligase Regulatory Subunit Glutamic acid decarboxylase; GAD Glutamic-oxaloacetic transaminase 2, mitochondrial Glutathione S Transferase Mu3 Glutathione S-Transferase Mu1 Glutathione S-Transferase Mu2 Glutathione S-Transferase Pi 1 Peptide 1 glutathione-S-transferase Glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) glycolipid; epitope is an alpha-galactose lactoseries carbohydrate glycoprotein, Cladosporium herbarum race 4 glycoprotein, Ulocladium glyoxylate cycle protein GOBP1 olfactory binding protein GOBP2 olfactory binding protein Golgi microtubule-associated protein Golgin-245 ortholog gp135/podocalyxin gp210 GPIb; von Willebrand factor receptor; GPIb/IX complex Grainyhead-like 1 Grainyhead-like 1 homolog Grainyhead-like protein 1 GRAM domain containing 4 Green fluorescent protein (GFP) Green fluorescent protein (GFP) variant Groucho protein Growth Factor Receptor-Bound Protein 2 Peptide 1 Growth factor receptor-bound protein 7 GST fused to a fragment of the Klarsicht alpha protein (amino acids 1895-2262) from Drosophila melanogaster GST fused to a fragment of the Klarsicht alpha protein (amino acids 277-556) from Drosophila melanogaster GST fused to a fragment of the Klarsicht alpha protein (amino acids 875-1169) from Drosophila melanogaster GTF2F2; General transcription factor IIF, polypeptide 2, 30kDa GTF2H2 family member C Guanine nucleotide binding protein (G protein), gamma 2 Gurken protein H6 family homeobox 3 Hairy/ enhancer-of-split related with YRPW notif protein Hairy/ enhancer-of-split related with YRPW notif protein 2 Half pint protein, C-terminal half (amino acids 277-637) HAND2; Heart and neural crest derivatives expressed 2 HCP4 HCS-1 (Hair Cell Soma-1) / otoferlin Headcase protein (amino acids 1-417) Heart and neural crest derivatives expressed 1 Heat shock 22kDa protein 8 Heat shock 27kDa protein 1 Heat Shock 27kDa Protein 1 Peptide 1 Heat Shock 27kDa Protein 1 Peptide 3 Heat shock 60kDa protein 1 (chaperonin) Heat Shock Protein-100 Heat shock transcription factor 2 Heat shock transcription factors 2 hemapoietic stem cell protein, 74 kDa Hematopoietic SH2 domain containing hemocytes, moth larvae heparan sulfate proteoglycan heparan sulfate proteoglycan (basement membrane) Hepatocyte nuclear factor 3-beta Hepatocyte nuclear factor 4, alpha Hepatoma derived growth factor-like 1 Hepatoma-derived growth factor Hepatoma-derived growth factor-related protein 2 HES1; Hes family bHLH transcription factor 1 HES3; Hes family bHLH transcription factor 3 HESX1; HESX homeobox 1 Heterochromatin Protein 1 hevin (SC1, SPARC-like 1) hexokinase (Type I isozyme) HHEX; Hematopoietically expressed homeobox HIF3A; Hypoxia inducible factor 3, alpha subunit High mobility group 20A High mobility group box 1 highwire hindsight: zinc-finger nuclear protein hindsight; Hisactophilin, Dd gelation factor histone 2A variant (gamma-H2AV) peptide phosphorylated (QDPQRKGNVILSQAY) Histone chaperone Anti-silencing function 1B Histone deacetylase 7 Histone deacetylase 8 Histone H4 Transcription Factors Histone lysine demethylase PHF8; PHD finger protein 8 Histone-arginine methyltransferase CARM1 Histone-lysine N-methyltransferase EZH2 Histone-lysine N-methyltransferase NSD2 Histone-lysine N-methyltransferase SETD7 Histone-lysine N-methyltransferase SMYD3 HLF; Hepatic leukemia factor HLFA, beta-subunit HMGA1; High mobility group AT-hook 1 HMGA1; High mobility group AT-hook 1 HMGB1; High mobility group box 1 HMGXB4; HMG box domain containing 4 HMR-1, amino acids 1099-1223 HNF3b / FoxA2 HNK-1 epitope Homeobox and leucine zipper encoding Homeobox and leucine zipper protein Homez Homeobox B2; HOXB2 Homeobox B6 Homeobox C8 Homeobox protein HMX3 Homeobox protein Hox- B5 Homeobox protein Meis 2 Homeobox protein Meis 3 Homeobox protein Nkx-2.5 Homeobox protein Nkx-3.2 Homeobox protein Nkx-6.1; Nkx6.1 Homeobox protein PKNOX2 Homeodomain-only protein HOP homeobox Hoxb4 Hoxc10 Hoxc9 HOXD9; Homeobox D9 HSF1; Heat shock transcription factor 1 HSF5; Heat shock transcription factor family member 5 HSP-60, amino acids 1-547 htsRC huntingtin Hydroxysteroid (17-beta) dehydrogenase 10 hypodermal marker hypodermis (seam cells) Ia antigen (chicken) Ia antigen (quail) ICAM-1 ID2; Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein ID3; Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein ID4; Inhibitor of DNA binding 4, dominant negative helix-loop-helix protein ID4; Inhibitor of DNA binding 4, dominant negative HLH protein IL2-inducible T-cell tyrosine-protein kinase InaD-like protein Indoleamine-pyrrole 2,3 dioxygenase Inhibitor of growth family, member 1 Inhibitor of growth family, member 4 INSM1; Insulinoma-associated 1 insulin (pro-) non-processed insulin (proinsulin; C-peptide) Insulin gene enhancer protein ISL-1 Insulin-like growth factor binding protein 2 integrin alpha 2 (CD49b, VLA2, ITGA2, ECMR II) integrin alpha V subunit (CD51, fibronectin receptor alpha subunit) integrin alpha V/5 + beta 5 (CD49e, CD51, fibronectin/vitronectin receptor) integrin alpha-2 integrin alpha-2 (CD49b) integrin alpha-2; alpha2beta1 integrin complex integrin alpha-2b beta-3 integrin alpha-3 integrin alpha-3 (CD49c) integrin alpha-4 (CD49d) integrin alpha-5 integrin alpha-5 (avian) integrin alpha-5 (CD49e) integrin alpha-6 integrin alpha-6 (avian) integrin alpha-7 (avian) integrin alphaPS1 (multiple edematous wing) integrin alphaPS2 (inflated) integrin beta integrin beta 3 (amino acids 26-40) integrin beta-1 integrin beta-1 (avian) integrin beta-1 (CD29) integrin beta-1 (Xenopus) integrin beta-2 (CD18) integrin beta-3 integrin betaPS (myospheroid) integrin, alpha-V beta-5 heterodimer Integrin, beta-1 subunit Integrin-linked protein kinase intercostal nerves intercostal nerves in a rostrocaudal gradient Interferon regulatory factor 3 Interferon regulatory factor 8 Interferon-inducible double stranded RNA dependent protein kinase activator A Interleukin 18 Interleukin 18 Peptide 1 Interleukin 6 intermediate filament subunit Intestine-specific Intraflagellar transport protein p139 Intraflagellar transport protein p172 Intraflagellar transport protein p57/55 Intraflagellar transport protein p81 IRF2; Interferon regulatory factor 2 IRF3; Interferon regulatory factor 3 Iroquois-class homeodomain protein IRX-5IRX5 ISL1; ISL LIM homeobox 1 Islet delta cells Islet-1 homeobox Islet-1 Islet-2 homeobox Islet-1 specific homeobox Islet-2 itchy homolog E3 ubiquitin protein ligase Jade -1 protein Jagged1 (JAG1) JAZF zinc finger 1 JAZF zinc finger 1; JAZF1 JAZF1; JAZF zinc finger 1 Junctions, epidermal cell:cell JUP; Junction plakoglobin KDM1A; Lysine (K)-specific demethylase 1A KDM8; Lysine (K)-specific demethylase 8 kel 1B Kelch Domain Containing 8A Peptide 1 keratan sulfate keratan sulfate (brain) keratin type I Keratin, type I cytoskeletal 19 Keratin, type II cytoskeletal 8; Cytokeratin endoA keratinocyte, basal, cell attachment antigen (human) KH domain containing, RNA binding, signal transduction associated 1 KH domain containing, RNA binding, signal transduction associated 3 KI-67; Marker of proliferation Ki-67 KIAA1967 kinesin Kinesin family member 13B Kinesin-like protein KIF2C Kinetochore protein NDC80 homolog KLF10; Kruppel-like factor 10 KLF10; Kruppel-like factor 10 KLF15; Kruppel-like factor 15 KLF16; Kruppel-like factor 16 KLOTHO Krueppel-like factor 15 Kruppel-like factor 1 (erythroid) Kruppel-like factor 10 Kruppel-like factor 14 Kruppel-like factor 17 Kruppel-like factor 5 (intestinal) Kruppel-like factor 7 L1 protein (rat) L1CAM and NCAM L1-like CAM (avian) L3MBTL4; L(3)mbt-like 4 (Drosophila) lactadherin lactase-phlorizin hydrolase Lactoylglutathione lyase lacunin lamin lamin A/C (recombinant) lamin C lamin Dm0 laminin laminin 5 (epiligrin, LAMA3, laminin-5 alpha subunit) laminin B2 laminin, gamma-1 chain laminin, S- laminin/fibronectin receptor laminin-binding lectin LAMP (gene symbol: Lsamp) LAMP-1 LAMP-1 (CHO) LAMP1; Lysosome-associated membrane glycoprotein 1 LAMP-2 LAMP-2 (110 kDa lysosomal membrane glycoprotein) LAMP-2 (CHO) LAMP-2 (human) late bloomer L-CAM, E-cadherin, uvomorulin LCOR; Ligand-dependent nuclear receptor corepressor LET-413 (amino acids 460-606) Lethal(3)malignant brain tumor-like protein 4 Leucine aminopeptidase Leucine rich repeat (in FLII) interacting protein 1 leukocyte, activated, cell surface glycoprotein LEUTX; Leucine twenty homeobox Ligand-dependent nuclear receptor corepressor-like Lim 1+2 / LhxV5 LIM domain binding 1 LIM homeobox 2; LHX2 LIM homeobox 4 LIM/homeobox protein Lhx2 Lim3 LIN-28 homolog A Lin-7 homolog C link protein lipooligosaccharide (LOS) LMP-1, amino acids 18-160 LMX L-myc protein lost protein, recombinant, expressed in E. coli lozenge Lymphoid-specific helicase, Lysine (K)-specific demethylase 2A Lysine (K)-specific demethylase 4A Lysine (K)-specific demethylase 4C Lysine (K)-specific demethylase 4E Lysine (K)-specific demethylase 6B lysine decarboxylase (E. corrodens) Lysine(K)-specific demethylase 6B Lysine-specific demethylase 2A lysosomal membrane glycoprotein (formerly called cv24) lysosomal membrane glycoprotein (LEP-100) / CD107 Lysosomal-associated membrane protein 1; Lamp-1 ; CD107a M2, membrane protein M6, membrane protein MAFB; V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B MAFF; V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F MAFG transcription factor major sperm protein (MSP) Malate dehydrogenase 1 NAD (soluble) Malate dehydrogenase 2, NAD (mitochondrial) maltase-glucoamylase Maltose binding protein mannose 6-phosphate / IGF II receptor, cation-independent mannose 6-phosphate receptor, cation-dependent MAT2B; Methionine adenosyltransferase II, beta Math1 (Atoh1) MAX; MYC associated factor X MAX-like protein X MAZ; MYC-associated zinc finger protein (purine-binding transcription factor) MBNL1 (muscleblind-like splicing regulator 1) MBNL2 protein (muscleblind-like splicing regulator 2) MBNL3 protein (muscleblind-like splicing regulator 3) MBP-sigma 1s MBP-sigma NS-His mCherry fluorescent protein MED4; Mediator complex subunit 4 Mediator complex subunit 1 Mediator of DNA-damage checkpoint 1 Megakaryocyte-associated tyrosine kinase Meis homeobox 2 Meis homeobox 3 MEIS2; Meis homeobox 2 melanoblasts and melanocytes of neural crest origin (avian) / glutathione S-transferase (alpha subunit) Melanoma Antigen Family A, 4 Menin Mesoderm posterior 1 homolog mesoderm, early, rodent Metastasin 100 calcium-binding protein A4 (Calvasculin) Methyl CpG Binding Protein 1 Methyl-CpG binding domain protein 4 METTL21A; Methyltransferase like 21A MHC class I MHC class II Microcephalin microfibrils (sea urchin) mineralocorticoid receptor Mitogen-activated protein kinase 1 Mitogen-activated protein kinase 14 Mitogen-activated protein kinase 8 Mitogen-activated protein kinase kinase kinase 7 Mitotic spindle assembly checkpoint protein MAD2A MLLT10; Protein AF-10 MLLT3; AF-9 MLX interacting protein Mmp1 catalytic domain Mmp1 hemopexin domain MNR2/HB9/Mnx1 Moesin Mortality factor 4-like 1 Mothers against decapentaplegic homolog 1 Mothers against decapentaplegic homolog 4 mouse Rb, last 200 amino acids (GST-Rb) MSH3 aa 24-308 MSX1+2 MUC17 CRD1 L CRD2 Mucin 1, cell surface associated Peptide 1 Mucin 1, cell surface associated Peptide 2 Multiple endocrine neoplasia I Muscle muscle fast C-protein muscle marker muscle slow C-protein muscle/neurite marker Muscleblind-like splicing regulator 3 Mutanase G MYC induced nuclear antigen |
MYCN; N-myc proto-oncogene protein Myeloid zinc finger 1 MYO1B; Myosin IB myoblast marker (avian) myoblast, chondrocyte marker (avian) myoblast, fibroblast marker (avian) Myocyte enhancer factor 2A Myocyte enhancer factor 2B Myocyte enhancer factor 2C MyoD Myogenin myomesin myosin (embryonic) myosin (human neonatal slow and fast IIa fibers) Myosin all isoforms myosin fast muscle light chain 2 myosin heavy chain Myosin heavy chain (all but 2X) Myosin heavy chain (all fast isoforms) Myosin heavy chain (human fast fibers) Myosin heavy chain (human nascent secondary and all fast fibers) Myosin heavy chain (human neonatal and adult fast fibers) Myosin heavy chain (human neonatal fast IIa fibers) Myosin heavy chain (human slow fibers) Myosin heavy chain (slow and 2A) Myosin heavy chain (slow, alpha- and beta-) myosin heavy chain A Myosin Heavy Chain Type I Myosin Heavy Chain Type IIA Myosin Heavy Chain Type IIB myosin heavy chain, adult myosin heavy chain, avian fast isoforms and rat, rabbit isoforms including type IIb fibers myosin heavy chain, avian fast isoforms and rat, rabbit, and turtle isoforms myosin heavy chain, avian fast isoforms and rat, turtle, newt, frog, fish, ray, and shark isoforms myosin heavy chain, avian slow isoforms; reacts with isoforms of other species myosin heavy chain, avian slow myosin and mammalian isoforms myosin heavy chain, avian slow myosin heavy chain 1, 2 and 3 myosin heavy chain, avian slow myosin heavy chain 2 and 3; slow muscle fibers in zebrafish myosin heavy chain, embryonic myosin heavy chain, embryonic and adult fast myosin heavy chain, embryonic and neonatal fast myosin heavy chain, fast, 2A myosin heavy chain, fast, 2B Myosin heavy chain, fast, 2X myosin heavy chain, fast, extraocular specific myosin heavy chain, fast, jaw muscle specific myosin heavy chain, multiple avian and rat, rabbit, turtle, newt and fish isoforms myosin heavy chain, neonatal and adult myosin heavy chain, neonatal fast myosin heavy chain, sarcomere myosin heavy chain, slow, SM1 only myosin heavy chain, slow, SM2 myosin heavy chain, slow, SM2 only myosin heavy chain, SM2 and atrial myosin heavy chain, ventricular myosin II myosin II heavy chain myosin IIB, cytoplasmic non-muscle myosin light chain 1 and 3f (LC1f/3f) myosin light chain 1s (LC1s) myosin light chain 1s, 2s, 1f and 2f (LC1s, LC1f, LC2s, LC2f) myosin, sarcomere (MHC) myosin-VIIa myotactin myotendinous antigen (tenascin) MZF1; Myeloid zinc finger 1 Na-K-Cl cotransporter Na-K-Cl cotransporters (shark) NAPA-73 (neurofilament-associated protein, 73 kDa) NCAM NCAM (cytoplasmic domain) NCAM (extracellular domain) NCAM (sialylated form); PSA-NCAM NCAM/L1CAM leech homologue NCK adaptor protein 1 NCOA1; Nuclear receptor coactivator 1 NEDD8-conjugating enzyme Ubc12 Nervana protein nervous system Nestin (intermediate filament protein) Nestin/radial glial marker neural associated ganglioside neural crest cells neural marker neural precursor cell expressed, developmentally down-regulated 4 isoform 1 neural precursor cells neural retinal gangliosides (9-0-acetyl-GD3) neural specific neural tube, dorsal neurocan (C-terminal epitope) neurocan (N-terminal epitope) neurocan receptor neurofilament (160 kDa) Neurofilament (165 kDa) Neurofilament associated neurofilaments neurofilaments, primary sensory and motor neurogenin 3 neuroglian (Drosophila, neuron-specific) neuroglian (Manduca sexta) Neuromedin-B peptide neuromuscular junction and reactive Schwann cell associated antigen neuron (motor) antigen neuronal cell surface marker neuronal cell surface marker (SC-1, DM-GRASP, BEN) neuronal marker (cytoplasmic) neuronal marker (TAG-1) neuronal, mesencephalic trigeminal cell surface marker neuronal, motor, marker (SC-1) Neurons, cephalic ganglia and ventral cord neurons, cytoplasmic neuropil region and primary motor neuron axons Neuropilin-1 neurotactin (Drosophila) NFATc1 NFATc2 NFE2L2; Nuclear factor, erythroid 2-like 2 NFKB1; Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 NFKB2; Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) NFYB; Nuclear transcription factor Y, beta NFYC; Nuclear transcription factor Y, subunit gamma NHE3, cation proton antiporter 3 nicotinic receptor, neuronal nidogen/entactin NINJ2; Ninjurin 2 Nkx2.2 transcription factor NKX2-5; NK2 homeobox 5 NKX3-1; NK3 homeobox 1 Nkx6.1 N-lysine methyltransferase SETD6 N-lysine methyltransferase SMYD2 N-myc (and STAT) interactor N-myc proto-oncogene protein NO66; Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66 Non-histone chrmosomal protein HMG-14. Non-histone chromosomal protein HMG-14 Norovirus RNA-dependent RNA polymerase Notch, extracellular domain, EGF repeats #12-20 Notch, extracellular domain, EGF repeats #5-7 Notch, intracellular domain Notch1 Notch2 notochord and neuropil notochord marker Nov NR5A1; Nuclear receptor subfamily 5, group A, member 1 NrCAM NRF1; Nuclear respiratory factor 1 Nuclear factor 1 A-type Nuclear factor I A-type Nuclear factor of activated T-cells, cytoplasmic 3 Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 2 Peptide 1 Nuclear factor, interleukin 3 regulated nuclear lamins II/III (Xenopus) nuclear membrane marker Nuclear receptor co-repressor 1 Nuclear receptor subfamily 0, group B, member 1 Nuclear receptor subfamily 1, group D, member 1 Nuclear receptor subfamily 1, group D, member 2 Nuclear receptor subfamily 2, group C, member 1 Nuclear receptor subfamily 5, group A, member 2 Nuclear respiratory factor 1 nucleolar protein nucleolin (95 kDa and 90 kDa isoforms) nucleolin (95 kDa isoform) nucleoplasmin Nucleoplasmin-3 Nucleoside Diphosphate Kinase A (nm23-H1) Nucleoside Diphosphate Kinase B Nucleosome assembly protein 1-like 5 Nucleus accumbens-accociated protein 1 Nucleus accumbens-associated 1 Nucleus accumbens-associated protein 2 Nucleus, intestine and epidermis nullo-GST fusion protein, an inframe of the entire nullo protein to c-terminus of GST Numb OBFC1; Oligonucleotide/oligosaccharide-binding fold containing 1 oligodendrocyte (myelin) marker (amphibian) oligodendrocytes and their processes oncomodulin (beta-parvalbumin) One cut homeobox 2 ONECUT1; One cut homeobox 1 orb protein orb2 protein ORC-2 Origin recognition complex, subunit 1 Ornithine Decarboxylase 1 Orthodenticle homeobox 2 Osa osteonectin osteopontin OTU domain, ubiquitin aldehyde binding 1 OTU domain, ubiquitin aldehyde binding 2 OTU domain-containing protein 6B Otx1 outer membrane protein-19 (OMP-1g) P granule P granule + body muscle P2X7 p46 cell surface protein excluded from macropinocytic cup p53 P6 protein N-terminus Haemophilus influenzae P6 surface antigen from Haemophilus influenzae p80 endosomal membrane protein P85A PABA peptide hydrolase (meprin) Paired box 5 Paired box 9 Paired related homeobox 2 PAR-3 paramyosin Parvimonas micra PAS-7 Patched PAWR; PRKC apoptosis WT1 regulator Pax3 Pax4 fusion protein (aa 132-171) PAX5; Paired box 5 Pax6 Pax7 PAX-interacting protein 1 PBP2 olfactory binding protein PBP3 olfactory binding protein PBX4; Pre-B-cell leukemia homeobox 4 PBX4; Pre-B-cell leukemia homeobox 4 Pdx1 PDZ and LIM Domain 1 peanut gene protein product PECAM Peptide II B 80kD Peregrin; Bromodomain and PHD finger containing, 1 Pericardin Perilipin 2 perlecan perlecan (domain IV) Peroxiredoxin 2 Peptide 1 Peroxiredoxin 2 Peptide 2 Peroxiredoxin 4 Peroxiredoxin 4 Peptide 1 Peroxisome proliferator-activated receptor alpha Peroxisome proliferator-activated receptor delta PGBD1; PiggyBac transposable element derived 1 Pgp-1 (Ly-24) lymphocyte cell adhesion glycoprotein pharyngeal marker PHD finger protein 10 PHD finger protein 19 PHD finger protein 21B PHD finger protein 7 PHD finger protein 8; Histone lysine demethylase PHF8 Phenylethanolamine N-methyltransferase phosphacan/protein tyrosine phosphatase – z/b Phosphatidylethanolamine Binding Protein 1 Phosphatidylinositol 3-kinase regulatory subunit alpha phosphatidylinositol-specific phospholipase C Phosphogluconate dehydrogenase Phosphoglycerate dehydrogenase Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) Phosphoinositide-3-kinase, regulatory subunit 2 (beta) Phosphoserine Aminotransferase 1 Phosphoserine phosphatase-like photoreceptors (rods and cones) photoreceptors (rods only) pigment cell marker (sea urchin) Pigment dispersing factor (PDF) precursor-related peptide Pigment-dispersing factor neuropeptide PIKFYVE PIKFYVE; Phosphoinositide kinase, FYVE finger containing PLAG1; Pleiomorphic adenoma gene 1 plasmalemma vesicle-associated protein (PLVAP) plateins (alpha-, beta- & gamma-) plateins (beta- & gamma-) PLXNB1; Plexin B1 Podoplanin POLE3; Polymerase (DNA directed), epsilon 3, accessory subunit Poly (ADP-ribose) polymerase 1 Poly (ADP-ribose) polymerase family, member 11 Polychaetoid Polycomb group ring finger 3 Polycomb group ring finger 6 Polymerase (DNA directed), lambda Polymerase (DNA directed), mu Pop1 (BVES) porin Porphyromonas gingivalis Porphyromonas gingivalis (cell surface antigen) Posterior sex combs protein Potassium channel modulatory factor 1 POU class 5, homeobox 1 POU class 6 homeobox 1 POU3F2; POU class 3 homeobox 2 POU5F1; POU class 5 homeobox 1 PPARD; Peroxisome proliferator-activated receptor delta PPP1R10; Protein phosphatase 1, regulatory subunit 10 PPP1R10; Serine/threonine-protein phosphatase 1 regulatory subunit 10 PPP5C; Protein phosphatase 5, catalytic subunit PR domain containing 7; PRDM7 PR domain zinc finger protein 1 PR domain zinc finger protein 13 PR domain zinc finger protein 4 PR domain zinc finger protein 8 PRDM16; PR domain containing 16 PRDM2; PR domain containing 2, with ZNF domain PRDM7; PR domain containing 7 PRDM7; PR domain containing 7 PRDM8; PR domain zinc finger protein 8 Pre-B-cell leukemia homeobox 1 Prevotella intermedia (cell surface antigen) Prevotella intermedia and nigrescens (cell surface antigen) primordial germ cell surface marker (mouse) PRMT7; Protein arginine methyltransferase 7 profilin profilin II proteins Proliferating Cell Nuclear Antigen Peptide 1 Prospero homeobox 1 Prospero protein Proteasome subunit Proteasome subunit 5 Protein arginine methyltransferase 6 Protein C-ets-1 Protein L-Myc Protein Phosphatase 2A Protein Smaug homolog 2 Protein tyrosine kinase 6 protein tyrosine phosphatase 69D Protein tyrosine phosphatase, non-receptor type 1 protein tyrosine phosphatase, non-receptor type 6 protein tyrosine phosphatase, receptor-linked, DPTP10D protein tyrosine phosphatase, receptor-linked, DPTP99A proteoglycan, hyaluronic acid binding region; aggrecan Proto-oncogene c-Fos Proto-oncogene tyrosine-protein kinase Src PRRX2; Paired related homeobox 2 PTP-ER Pumilio homolog 1 puromycin Putative histone-lysine N-methyltransferase PRDM6 Quail cell marker Quail endothelial cell surface RAC-gamma serine/threonine-protein kinase RAD18 Homolog Peptide 1 RAD52 homolog (S. cerevisiae) radial glial cells radial glial cells / vimentin Rarres2 Ras-related C3 botulinum toxin substrate 1 (rho family small GTP binding protein Rac1) RBBP5; Retinoblastoma binding protein 5 RCOR2; REST corepressor 2 RecD protein Receptor-type tyrosine-protein phosphatase delta Receptor-type tyrosine-protein phosphatase epsilon Receptor-type tyrosine-protein phosphatase-like N reelin Reg1a Reg3g Regulation of nuclear pre-mRNA domain containing 1A RelA-associated inhibitor relaxin hormone Relish reovirus reovirus capsid protein mu 1C reovirus gamma 2 vertex protein reovirus outer capsid protein sigma 3 reovirus sigma 1s reovirus sigma NS Repo; Reversed polarity protein REST corepressor 3 Retina and anterior neural fold homeobox Retinal homeobox protein Rx retinal space (mechanoreceptors?) Retinoic acid receptor RXR-gamma Retinoic acid receptor, beta Retinoid X receptor RXR- gamma Retinoid X receptor, gamma RFX5; DNA-binding protein RFX5 RFX6; Regulatory factor X, 6 RFXANK; Regulatory factor X-associated ankyrin-containing protein RGCC; Regulator of cell cycle RGS12 (“Regulator of G Protein Signaling-12”); N-terminal peptide PDZ & PTB domains Rho guanine nucleotide exchange factor (GEF) 12 rho1 rhoB rhodopsin 1 (Rh1 Drosophila) Rhox homeobox family, member 2 Ribonucleotide Reductase M2 Peptide 1 Ribonucleotide Reductase M2 Peptide 2 ribosomal protein L3 Ribosomal protein S6 kinase alpha-5 ribosome, (E. coli) ribosome, 30S (E. coli) rim RME-1, amino acids 333-575 RNA binding protein 8A Rnase1 RNF113B; Ring finger protein 113B RNF20; Ring finger protein 20, E3 ubiquitin protein ligase Robo (Drosophila) Robo1 (vertebrate) Robo3 cytoplasmic (Drosophila) Robo3 extracellular (Drosophila) Rop Ror2 (Nt 2535-2835) RORA; RAR-related orphan receptor A RORgt rough Drosophila protein Rumpelstiltskin Runt-related transcription factor 2 RUVBL1; RuvB-like AAA ATPase 1 RXRA; Retinoid X receptor, alpha ryanodine receptor, skeletal ryanodine receptors S100 calcium binding protein A2 SAC1 S-adenosylhomocysteine hydrolase sarcalumenin SATB1; SATB homeobox 1 SAX7, recombinant amino acids 1051-1144 scabrous Drosophila gene protein SCAND3; SCAN domain containing 3 SCAR Schwann cell & myoblast plasma membrane glycoprotein Schwann cell marker, P(o) (avian) Schwann cell myelin protein Scm-like with four mbt domains 1 Scm-like with four MBT domains 2 SCRT1; Scratch family zinc finger 1 Secretory cell, peripharyngeal selectin, E- Selenoprotein P Sema I (grasshopper) Sema II sensory cilia and excretory pore marker Serine/threoinine-protein kianse Chk2 Serine/threonine-protein kinase Chk2; Checkpoint kinase 2 Serine/threonine-protein kinase D1 Serine/threonine-protein kinase OSR1 Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform Serpin Peptidase Inhibitor, Clade A (Alpha-1 Antiproteinase, Antitrypsin) Member 6 Peptide 1 Serpin Peptidase Inhibitor, Clade A, Member 1 Serrate, RNA effector molecule SET nuclear proto-oncogene Seven-up Sex comb on midleg-like protein 4 Sex combs reduced homeotic protein Sex determining region Y protein sex-lethal protein SH2 domain containing 1A; Duncan disease SH2- protein; SLAM-associated protein SH2 domain containing 4A SH2 domain-containing protein 1A SH2 domain-containing protein 3A SH2 domain-containing protein 4A SH2B adaptor protein 2 SH3-domain binding protein 2 Sharpin SHC (Src homology 2 domain containing) family, member 4 Shot sialomucin complex (Muc4) sidestep Signal transducer and activator of transcription 1, 91kDa Signal transducer and activator of transcription 1-alpha/beta Signal transducer and activator of transcription 4 Signal transducer and activator of transcription 5A Signal transducer and activator of transcription 5B Sir2 (Drosophila) SIRT2; Sirtuin 2 Sirtuin 1 Sirtuin 2 Sirtuin 5, mitochondrial Sirtuin 5; SIX homeobox 5 SIX5 skeletal muscle marker, 102 kDa SKI-like protein SLC25A24; Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 SLC30A9; Zinc transporter 9 SLC4A1, anion exchanger 1 slit protein, C-terminal Slug SMAD family member 3 SMAD family member 7 Smad nuclear-interacting protein 1 SMAD4; SMAD family member 4 Small ubiquitin-related modifier (SUMO) SMARCC2; SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 SMARCE1; SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 SMN protein aa 159-209 SMN protein aa 159-209 Exon 4 SMN protein aa 210-241 Exon 5 SMN protein aa 28-91 SMN-interacting protein-1 (Gemin2) Smoothened (Smo) Snail family zinc finger 3 snRNA-activating protein complex subunit 4 SNW domain containing 1 Sodium/bile acid cotransporter Solute Carrier Family 31 (Copper Transporter), Member 1 Peptide 1 Solute Carrier Family 31 (Copper Transporter), Member 2 Peptide 1 somatomedin-C; Sm-C/IGF-1 sonic hedgehog SOX12; SRY (sex determining region Y)-box 12 SOX3; Transcription factor SOX-3 SOX4; (SRY sex determining region Y)-box 4 SOX4; SRY (sex determining region Y)-box 4 SP110 nuclear body protein SP140 nuclear body protein SPARC (osteonectin) SPARC, recombinant human Spectrin, alpha Spermatogenesis and oogenesis specific basic helix-loop-helix 1 Spermidine or Spermine N1-Acetyltransferase 1 Spindle-F full length protein Spitz, extracellular domain Spleen tyrosine kinase Squamous cell carcinoma antigen 1 Squash protein, CG 4711, full length protein Squid A protein, full length Squid S protein, full length SQV-8, amino acids 150-349 Src homology 2 domain containing transforming protein D SRY (sex determining region Y)-box 13 SRY (sex determining region Y)-box 4 SRY (sex determining region Y)-box 9 SSBP4; Single stranded DNA binding protein 4 SSEA-1/ alpha-(1,3)-fucosyltransferase / CD15 SSEA-3 (carbohydrate from 4-8 cell mouse embryos) SSEA-4 (carbohydrate from human embryonal carcinoma) STAM-binding protein STAT1; Signal transducer and activator of transcription 1, 91kDa STAT2; Signal transducer and activator of transcription 2, 113kDa STAT3; Signal transducer and activator of transcription 3 STAT4; Signal transducer and activator of transcription 4 STAT5A; Signal transducer and activator of transcription 5A STAT5B; Signal transducer and activator of transcription 5B Steroidogenic factor 1 stromal cell surface marker sucrase-isomaltase Sulfotransferase family 1E, estrogen-preferring, member 1 [Homo sapiens] SUMO aa76-86 SUMO-1 SUMO-2 Suppressor of cytokine signaling 2 Suppressor of fused (Su (fu)) SWI/SNF complex subunit SMARCC1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related SYAP1; Synapse associated protein 1 sympathoadrenal marker Synapse-associated protein 47 kDa; SAP47 Synapsin Synaptic vesicle glycoprotein 2A; synaptic vesicles synaptobrevin Synaptotagmin synaptotagmin 1, cytoplasmic domain synaptotagmin B synaptotagmin, cytoplasmic domain Sync Syntaxin synuclein, alpha & beta synuclein, gamma (breast cancer-specific protein 1) Peptide 1 synuclein-gamma TAC-1 TADA2B; Transcriptional adaptor 2B talin talin (avian) Talin carboxy terminus 534 amino acids talinA (N-terminal) Tango Tannerella forsythia TAR (HIV-1) RNA binding protein 2 Tau TAX1BP1; Tax1 (human T-cell leukemia virus type I) binding protein 1 T-box 19 T-box transcription factor TBX19 T-box transcription factor TBX4 TBX19; T-box 19 TBX4; T-box 4 TCF25 Transcription Factors 25 (basic helix-loop-helix) TEA domain family member 3 Telomeric Repeat Binding Factor 2 Peptide 1 tenascin Tensin 1 Tensin 3 Tensin 4 TERF2; Telomeric repeat binding factor 2 TERF2IP; Telomeric repeat binding factor 2, interacting protein tetraspanin large extracellular loop amino acids 111-217 TFAP2D; Transcription factor AP-2 delta TGFb3, active domain TGFb3, latent domain TGIF1; TGFB-induced factor homeobox 1 TGIF2LX; TGFB-induced factor homeobox 2-like, X-linked THAP domain containing 5 THAP domain containing 9 THAP domain-containing protein 11 THAP domain-containing protein 4 THAP10; THAP domain containing 10 Thyroid hormone receptor, alpha Thyroid Stimulating Hormone, Beta Peptide 1 Thyroid Stimulating Hormone, beta Peptide 2 Titin Topoisomerase I topoisomerase I mitochondrial TOX high mobility group box family member 3 TOX3; TOX high mobility group box family member 3 TP53; Tumor protein p53 Transcription elongation factor A (SII)-like 8 Transcription facotr RelB Transcription factor Sp5 Transcription factor 25 Transcription factor AP-2 alpha Transcription factor AP-2 beta Transcription factor AP-2 gamma Transcription factor CP2; Alpha-globin transcription factor CP2 Transcription factor elt-2 Transcription factor ETV6; Ets variant 6 Transcription factor Ovo-like 2 Transcription factor p65 or RELA Transcription factor RelB Transcription factor RFX3 Transcription factor SOX-9 Transcription factor Spi-C Transcription Factors 24 Transcription Factors 25 (basic helix-loop-helix) Transcription Factors AP-2 alpha (activating enhancer binding protein 2 alpha) transcription factors E2F 6 transcription factors Ovo-like 2 transcription factors Spi-C Transcriptional adaptor 3 Transcriptional enhancer factor TEF-3 Transcriptional regulator ERG Transcriptional repressor NF-X1 transferrin receptor Transgelin Peptide 1 transglutaminase, tissue transitin transketolase Transthyretin triadin Trio Tripartite motif containing 24 tRNA aspartic acid methyltransferase 1 tropomyosin tropomyosin, chicken gizzard tropomyosin, gizzard tropomyosin, muscle tropomyosin, recombinant human, isoform 5 and 4 fusion (hTM5/4) protein tropomyosin, recombinant, isoform 5 and 4 fusion (hTM5/4) protein troponin I, cardiac troponin I, skeletal and cardiac troponin T troponin T, cardiac troponin T, skeletal muscle specific TRP protein (transient receptor potential) truncated version of sry-a protein which contains amino acids 46 to 530 of sry-a TSC22 domain family, member 1 TSC22D4; TSC22 domain family, member 4 TSLP Tubulin tubulin (alpha-) tubulin (beta-) Tubulin, beta Tudor and KH domain containing Tumbleweed Tumor necrosis factor (ligand) superfamily, member 10 Tumor necrosis factor receptor superfamily member 9 Tumor protein p53 Tumor-associated Calcium Signal Transducer 2 tyrosine 3-monooxygenase or tryptophan 5-monooxygenase activation protein, beta polypeptide tyrosine 3-monooxygenase or tryptophan 5-monooxygenase activation protein, epsilon polypeptide tyrosine hydroxylase Tyrosine-protein kinase Fes/Feline sarcoma oncogene Tyrosine-protein kinase Fyn/ FYN oncogene related to SRC, FGR, YES Tyrosine-protein ZAP-70 Tyrosyl-DNA Phosphodiesterase 2 Ubiquitin carboxyl-terminal hydrolase 14 Ubiquitin carboxyl-terminal hydrolase 16 Ubiquitin carboxyl-terminal hydrolase isozyme L1 Ubiquitin carboxyl-terminal hydrolase L3 (ubiquitin thiolesterase) Ubiquitin carboxyl-terminal hydrolase L5 Ubiquitin Conjugating Enzyme E2C Peptide 1 Ubiquitin Conjugating Enzyme E2C Peptide 2 Ubiquitin Conjugating Enzyme E2E2 Ubiquitin Conjugating Enzyme E2L 6 Ubiquitin Conjugating Enzyme E2L 6; UBE2E1 Ubiquitin protein ligase E3A Ubiquitin specific peptidase 7 Ubiquitin-conjugating enzyme E2 A Ubiquitin-conjugating enzyme E2 B Ubiquitin-conjugating enzyme E2 D1 Ubiquitin-conjugating enzyme E2 Q1 ubiquitin-conjugating enzyme E2, J1 Ubiquitin-conjugating enzyme E2C ubiquitin-conjugating enzyme E2E 1 Ubiquitin-conjugating Enzymes E2B Ubiquitin-like modifier activating enzyme 1 Ubiquitin-protein ligase E3A UBQLN2; Ubiquilin 2 Ultrabithorax 5′ exon (107 amino acids) Ultrabithorax protein UNC-10, amino acids 1-144 Upstream transcription factor 2, c-fos interacting Us9 (pseudorabies virus) USF2; Upstream transcription factor 2, c-fos interacting utrophin utrophin aa 113-371 V_H_ATpase c-subunit vasa vasa protein Vav 1 guanine nucleotide exchange factor Vav 2 guanine nucleotide exchange factor VCAM VCAM-1 V-crk avian sarcoma virus CT10 oncogene homolog VENTX homeobox protein VENTX; VENT homeobox VENTX; VENT homeobox v-erb-b2 erythroblastic leukemia viral oncogene homolog Peptide 1 versican (hyaluronate-binding region) Villin-like protein quail; 6B9 Vimentin vimentin (Xenopus) vimentin; radial and radial glial cell marker vinculin vinculin, meta-vinculin visinin v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog Wash Wash cDNA/GST fusion protein Wasp WDHD1; WD repeat and HMG-box DNA binding protein 1 Whamy Windbeutel protein, N-terminal half Wingless protein wit Wolf-Hirschhorn syndrome candidate 1 wound epithelium & transport / secretory cytoskeletal protein wound epithelium & transport/secretory cell protein, 42 kDa wrapper Xenopus nuclear factor, xnf7 Xenotropic murine leukemia virus-related virus P12 X-prolyl aminopeptidase (aminopeptidase P) 1, soluble Peptide 1 X-prolyl aminopeptidase (aminopeptidase P) 1, soluble Peptide 2 Yan Drosophila protein YAP1 (YAP65 / Yes-associated protein 1) Y-box-binding protein 3 YBX1; Y box binding protein 1 YBX2; Y box binding protein 2 YEATS domain containing 4 YEATS domain-containing 4 YEATS2; YEATS domain containing 2 YY1 transcription repressor protein YY1 transcriptional repressor protein; Yin-Yang-1 YY2 Transcription Factors ZBTB16; Zinc finger and BTB domain containing 16 ZBTB2; Zinc finger and BTB domain containing 2 ZBTB25; Zinc finger and BTB domain containing 25 ZBTB5; Zinc finger and BTB domain containing 5 ZBTB5; Zinc finger and BTB domain containing 5 ZCCHC7; Zinc finger, CCHC domain containing 7 ZEB2; Zinc finger E-box binding homeobox 2 Zeta-chain (TCR) associated protein kinase 70kDa zeugmatin ZFP3 zinc finger protein ZFP41 zinc finger protein ZFP90; ZFP90 zinc finger protein ZFP92 zinc finger protein ZFYVE19; Zinc finger, FYVE domain containing 19 ZFYVE20; Zinc finger, FYVE domain containing 20 Zic family member 2 ZIC1; Zic family member 1 ZIM2; Zinc finger, imprinted 2 zinc alpha-2-glycoprotein (ZAG) Zinc finger and BTB domain containing 18 Zinc finger and BTB domain containing 21 Zinc finger and BTB domain containing 37 Zinc finger and BTB domain containing 39 Zinc finger and BTB domain containing 46 Zinc finger and BTB domain containing 7B Zinc finger and BTB domain containing 7C Zinc finger and BTB domain containing protein 32 Zinc finger and SCAN domain containing 2 Zinc finger and SCAN domain containing 25 Zinc finger and SCAN domain containing 5A Zinc finger and SCAN domain containing 9 Zinc finger and SCAN domain-containing protein 26 Zinc finger and SCAN domain-containing protein 29 Zinc finger CCCH domain-containing protein 15 Zinc finger CCCH-type containing 7A Zinc finger CCHC domain-containing protein 7 Zinc finger E-box binding homeobox 2 Zinc finger family member 783 Zinc finger protein 134 Zinc finger protein 140 Zinc finger protein 155 Zinc finger protein 157 Zinc finger protein 165 Zinc finger protein 169 Zinc finger protein 18 Zinc finger protein 189 Zinc finger protein 200 Zinc finger protein 202 Zinc finger protein 207 Zinc finger protein 222 Zinc finger protein 230 Zinc finger protein 239 Zinc finger protein 239; ZNF239 Zinc finger protein 24 Zinc finger protein 263 Zinc finger protein 264 Zinc finger protein 280A Zinc finger protein 280D Zinc finger protein 333 Zinc finger protein 341 Zinc finger protein 354A Zinc finger protein 354B Zinc finger protein 358 Zinc finger protein 384 Zinc finger protein 385B Zinc finger protein 397 Zinc finger protein 408 Zinc finger protein 410 Zinc finger protein 428 Zinc finger protein 438 Zinc finger protein 446 Zinc finger protein 467; ZNF467 Zinc finger protein 485 Zinc finger protein 496 Zinc finger protein 521 Zinc finger protein 526 Zinc finger protein 543 Zinc finger protein 548 Zinc finger protein 549 Zinc finger protein 550 Zinc finger protein 554 Zinc finger protein 574 Zinc finger protein 581 Zinc finger protein 583 Zinc finger protein 584 Zinc finger protein 597 Zinc finger protein 608 Zinc finger protein 623 Zinc finger protein 648 Zinc finger protein 654 Zinc finger protein 654; melanoma-associated antigen Zinc finger protein 683 Zinc finger protein 696 Zinc finger protein 697 Zinc finger protein 701 Zinc finger protein 75a Zinc finger protein 770 Zinc finger protein 784 Zinc finger protein 792 Zinc finger protein 8 Zinc finger protein 800 Zinc finger protein 81 Zinc finger protein Gfi-1 Zinc finger protein ZFAT Zinc finger protein ZFPM2 Zinc finger with KRAB and SCAN domains 7 Zinc finger with KRAB and SCAN domains 8 Zinc Finger, AN1-type Domain 6 Zinc finger, BED-type containing 1 Zinc finger, BED-type containing 3 Zinc finger, FYVE domain containing 20 (Rabenosyn-5) Zinc finger, imprinted 2 Zinc finger, matrin-type 2 Zinc finger, matrin-type 4 Zinc finger, MYM-type 3 Zinc finger, ZZ-type containing 3 Zinc fingers and homeoboxes 1 Zinc fingers and homeoboxes 2 Zinc fingers and homeoboxes 3 ZKSCAN1; Zinc finger with KRAB and SCAN domains 1 ZKSCAN2; Zinc finger with KRAB and SCAN domains 2 ZKSCAN3; Zinc finger with KRAB and SCAN domains 3 ZMYM3; Zinc finger, MYM-type 3 ZNF157; Zinc finger protein 157 ZNF165; Zinc finger protein 165 ZNF174; Zinc finger protein 174 ZNF175; Zinc finger protein 175 ZNF18; Zinc finger protein 18 ZNF192; Zinc finger with KRAB and SCAN domains 8 ZNF197; Zinc finger protein 197 ZNF202; Zinc finger protein 202 ZNF207; Zinc finger protein 207 ZNF212; Zinc finger protein 212 ZNF217; Zinc finger protein 217 ZNF23; Zinc finger protein 23 ZNF230; Zinc finger protein 230 ZNF232; Zinc finger protein 232 ZNF277; Zinc finger protein 277 ZNF296; Zinc finger protein 296 ZNF324; Zinc finger protein 324 ZNF324; Zinc finger protein 324 ZNF354A; Zinc finger protein 354A ZNF384; Zinc finger protein 384 ZNF397; Zinc finger protein 397 ZNF410; Zinc finger protein 410 ZNF444; Zinc finger protein 444 ZNF446; Zinc finger protein 446 ZNF48; Zinc finger protein 48 ZNF483; Zinc finger protein 483 ZNF488; Zinc finger protein 488 ZNF503-AS2; Zinc finger protein 503-AS2 ZNF556; Zinc finger protein 556 ZNF585B; Zinc finger protein 585B ZNF639; Zinc finger protein 639 ZNF689; Zinc finger protein 689 ZNF783; Zinc finger family member 783 ZNF784; Zinc finger protein 784 ZNF830; Zinc finger protein 830 ZNF846; Zinc finger protein 846 ZNFX1; Zinc finger, NFX1-type containing 1 ZO-1 tight junction protein ZSCAN12; Zinc finger and SCAN domain containing 12 ZSCAN2; Zinc finger and SCAN domain containing 2 ZSCAN21; Zinc finger and SCAN domain containing 21 ZSCAN22; Zinc finger and SCAN domain containing 22 ZSCAN23; Zinc finger and SCAN domain containing 23 ZSCAN31; Zinc finger and SCAN domain containing 31 ZSCAN5A; Zinc finger and SCAN domain containing 5A ZUFSP; Zinc finger with UFM1-specific peptidase domain Zw5 ZXD family zinc finger C ZXDC; ZXD family zinc finger C ZZZ3; Zinc finger, ZZ-type containing 3 |
Major histocompatibility complex
The major histocompatibility complex (MHC) is a set of cell surface proteins essential for the acquired immune system to recognize foreign molecules in vertebrates, which in turn determines histocompatibility. The main function of MHC molecules is to bind to peptide fragments derived from pathogens and display them on the cell surface for recognition by the appropriate T-cells. MHC molecules mediate interactions of leukocytes, also called white blood cells (WBCs), which are immune cells, with other leukocytes or with body cells. The MHC determines compatibility of donors for organ transplant, as well as one’s susceptibility to an autoimmune disease via crossreacting immunization. The human MHC is also called the HLA (human leukocyte antigen) complex (often just the HLA). The mouse MHC is called the H-2 complex or H-2.
In a cell, protein molecules of the host’s own phenotype or of other biologic entities are continually synthesized and degraded. Each MHC molecule on the cell surface displays a molecular fraction of a protein, called an epitope. The presented antigen can be either self or non-self, thus preventing an organism’s immune system targeting its own cells. In its entirety, the MHC population is like a meter indicating the balance of proteins within the cell.
The MHC gene family is divided into three subgroups: class I, class II, and class III. Class I MHC molecules have β2 subunits so can only be recognised by CD8 co-receptors. Class II MHC molecules have no β2 subunits so can be recognised by CD4 co-receptors. In this way MHC molecules chaperone which type of lymphocytes may bind to the given antigen with high affinity, since different lymphocytes express different TCR co-receptors.
Diversity of antigen presentation, mediated by MHC classes I and II, is attained in at least three ways: (1) an organism’s MHC repertoire is polygenic (via multiple, interacting genes); MHC expression is codominant (from both sets of inherited alleles); MHC gene variants are highly polymorphic (diversely varying from organism to organism within a species). Major histocompatibility complex and sexual selection has been observed in male mice making mate choices of females with different MHCs and thus demonstrating sexual selection. Also, at least for MHC I presentation, there has been evidence of antigenic peptide splicing which can combine peptides from different proteins, vastly increasing antigen diversity.
In immunity
Of the three MHC classes identified, attention commonly focuses on classes I and II. By interacting with CD4 molecules on surfaces of helper T cells, MHC class II mediates establishment of specific immunity (also called acquired immunity or adaptive immunity). By interacting with CD8 molecules on surfaces of cytotoxic T cells, MHC class I mediates destruction of infected or malignant host cells, the aspect of specific immunity termed cellular immunity. (The other arm of specific immunity is humoral immunity, whose relation to MHC is more indirect)
Functions
MHC is the tissue-antigen that allows the immune system (more specifically T cells) to bind to, recognize, and tolerate itself (autorecognition). MHC is also the chaperone for intracellular peptides that are complexed with MHCs and presented to TCRs as potential foreign antigens. MHC interacts with TCR and its co-receptors to optimize binding conditions for the TCR-antigen interaction, in terms of antigen binding affinity and specificity, and signal transduction effectiveness.
Essentially, the MHC-peptide complex is a complex of autoantigen/alloantigen. Upon binding, T cells should in principle tolerate the auto-antigen, but activate when exposed to the allo-antigen. Disease states occur when this principle is disrupted.
Antigen presentation: MHC molecules bind to both T cell receptor and CD4/CD8 co-receptors on T lymphocytes, and the antigen epitope held in the peptide-binding groove of the MHC molecule interacts with the variable Ig-Like domain of the TCR to trigger T-cell activation.
Autoimmune reaction: Having some MHC molecules increases the risk of autoimmune diseases more than having others. HLA-B27 is an example. It is unclear how exactly having the HLA-B27 tissue type increases the risk of ankylosing spondylitis and other associated inflammatory diseases, but mechanisms involving aberrant antigen presentation or T cell activation have been hypothesized.
Tissue allorecognition: MHC molecules in complex with peptide epitopes are essentially ligands for TCR. T cells become activated by binding to the peptide-binding grooves of any MHC molecule that T cells were not entrained to recognize during thymus positive selection.
Lymphocytes
As a lineage of leukocytes, lymphocytes reside in peripheral lymphoid tissues, including lymphoid follicles and lymph nodes, and include B cells, T cells, and natural killer cells (NK cells). B cells, which act specifically, secrete antibody molecules, but do not bind MHC. T cells, which act specifically, as well as NK cells, which act innately, interact with MHC. NK cells express Killer Ig-like receptors (KIRs) that bind to MHC I molecules and signal through ITIM (immunoreceptor tyrosine inhibition motif) recruitment and activation of protein tyrosine phosphatases. This means in contrast to CD8/TCR interaction that activates Tc lymphocytes, NK cells becomes deactivated when bound to MHC I. When MHC class I expression is low, as is typically the case with abnormal cell function during viral infection or tumourigenesis, NK cells lose the inhibitory KIR signal and trigger programmed cell death of the abnormal cell. NK cells thus help prevent progress of cancerous cells by contributing to tumor surveillance.
MHC class II
MHC class II can be conditionally expressed by all cell types, but normally occurs only on professional antigen-presenting cells (APCs): macrophages, B cells, and especially dendritic cells (DCs). An APC takes up an antigen, performs antigen processing, and returns a molecular fraction of it—a fraction termed the epitope—to the APC’s surface, coupled within an MHC class II molecule mediating antigen presentation by displaying this epitope. On the cell’s surface, the epitope can contact its cognate region on immunologic structures recognizing that epitope. That molecular region which binds to—or, in jargon, ligates—the epitope is the paratope.
On surfaces of helper T cells are CD4 receptors, as well as T cell receptors (TCRs). When a naive helper T cell’s CD4 molecule docks to an APC’s MHC class II molecule, its TCR can meet and be imprinted by the epitope coupled within the MHC class II. This event primes the naive helper T cell. According to the local milieu, that is, the balance of cytokines secreted by APCs in the microenvironment, the naive helper T cell (Th0) polarizes into either a memory Th cell or an effector Th cell of phenotype either type 1 (Th1), type 2 (Th2), type 17 (Th17), or regulatory/suppressor (Treg), as so far identified, the Th cell’s terminal differentiation.
MHC class II thus mediates immunization to—or, if APCs polarize Th0 cells principally to Treg cells, immune tolerance of—an antigen. The polarization during primary exposure to an antigen is key in determining a number chronic diseases, such as inflammatory bowel diseases and asthma, by skewing the immune response that memory Th cells coordinate when their memory recall is triggered upon secondary exposure to similar antigens. (B cells express MHC class II to present antigen to Th0, but when their B cell receptors bind matching epitopes, interactions which are not mediated by MHC, these activated B cells secrete soluble immunoglobulins: antibody molecules mediating humoral immunity.)
MHC class I
MHC class I occurs on all nucleated cells and also in platelets—in essence all cells but red blood cells. It presents epitopes to killer T cells, also called cytotoxic T lymphocytes (CTLs). A CTL expresses CD8 receptors, in addition to TCRs. When a CTL’s CD8 receptor docks to a MHC class I molecule, if the CTL’s TCR fits the epitope within the MHC class I molecule, the CTL triggers the cell to undergo programmed cell death by apoptosis. Thus, MHC class I helps mediate cellular immunity, a primary means to address intracellular pathogens, such as viruses and some bacteria, including bacterial L forms, bacterial genus Mycoplasma, and bacterial genus Rickettsia. In humans, MHC class I comprises HLA-A, HLA-B, and HLA-C molecules.
Genes
MHC gene families are found in all vertebrates, though they vary widely. In humans, the MHC region occurs on chromosome 6, between the flanking genetic markers MOG and COL11A2 (from 6p22.1 to 6p21.3 about 29Mb to 33Mb on the hg19 assembly), and contains 240 genes spanning 3.6 megabase pairs (3 600 000 bases).[8] About half have known immune functions.
The same markers in the gray short-tailed opossum (Monodelphis domestica), a marsupial, span 3.95 Mb, yielding 114 genes, 87 shared with humans. Marsupial MHC genotypic variation lies between eutherian mammals and birds, taken as minimal MHC encoding, but is closer in organization to that of nonmammals, and MHC class I genes of marsupials have amplified within the class II region, yielding a unique class I/II region.
Class III functions very differently from class I and class II, but its locus occurs between the other two classes, on chromosome 6 in humans, and are frequently discussed together.
| Class | Encoding | Expression |
| I | (1) peptide-binding proteins, which select short sequences of amino acids for antigen presentation, as well as (2) molecules aiding antigen-processing (such as TAP and tapasin). | One chain, called α, whose ligands are the CD8 receptor—borne notably by cytotoxic T cells—and inhibitory receptors borne by NK cells |
| II | (1) peptide-binding proteins and (2) proteins assisting antigen loading onto MHC class II’s peptide-binding proteins (such as MHC II DM, MHC II DQ, MHC II DR, and MHC II DP). | Two chains, called α & β, whose ligands are the CD4 receptors borne by helper T cells. |
| III | Other immune proteins, outside antigen processing and presentation, such as components of the complement cascade (e.g., C2, C4, factor B), the cytokines of immune signaling (e.g., TNF-α), and heat shock proteinsbuffering cells from stresses | Various |
Proteins
MHC proteins have immunoglobulin-like structure.
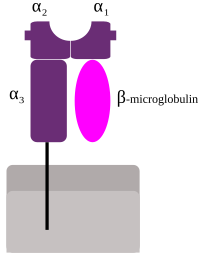
MHC class I protein molecule
MHC Class I
Classical MHC molecules present epitopes to the TCRs of CD8+ T lymphocytes. Nonclassical molecules (MHC class IB) exhibit limited polymorphism, expression patterns, and presented antigens; this group is subdivided into a group encoded within MHC loci (e.g., HLA-E, -F, -G), as well as those not (e.g., stress ligands such as ULBPs, Rae1, and H60); the antigen/ligand for many of these molecules remain unknown, but they can interact with each of CD8+ T cells, NKT cells, and NK cells.
 MHC Class II
MHC Class II
MHC class II is formed of two chains, α and β, each having two domains—α1 and α2 and β1 and β2—each chain having a transmembrane domain, α2 and β2, respectively, anchoring the MHC class II molecule to the cell membrane. The peptide-binding groove is formed of the heterodimer of α1 and β1.
MHC class II molecules in humans have five to six isotypes. Classic molecules present peptides to CD4+ lymphocytes. Nonclassic molecules, accessories, with intracellular functions, are not exposed on cell membranes, but in internal membranes in lysosomes, normally loading the antigenic peptides onto classic MHC class II molecules.
MHC Class III
Class III molecules have physiologic roles unlike classes I and II, but are encoded between them in the short arm of human chromosome 6. Class III molecules include several secreted proteins with immune functions: components of the complement system(such as C2, C4, and B factor), cytokines (such as TNF-α, LTA, and LTB), and heat shock proteins.
Antigen processing and presentation

MHC class I pathway: Proteins in the cytosol are degraded by the proteasome, liberating peptides internalized by TAPchannel in the endoplasmic reticulum, there associating with MHC-I molecules freshly synthesized. MHC-I/peptide complexes enter Golgi apparatus, are glycosylated, enter secretory vesicles, fuse with the cell membrane, and externalize on the cell membrane interacting with T lymphocytes.
Peptides are processed and presented by two classical pathways:
- In MHC class II, phagocytessuch as macrophages and immature dendritic cells take up entities by phagocytosisinto phagosomes—though B cells exhibit the more general endocytosis into endosomes—which fuse with lysosomes whose acidic enzymes cleave the uptaken protein into many different peptides. Via physicochemical dynamics in molecular interaction with the particular MHC class II variants borne by the host, encoded in the host’s genome, a particular peptide exhibits immunodominanceand loads onto MHC class II molecules. These are trafficked to and externalized on the cell surface.
- In MHC class I, any nucleated cell normally presents cytosolic peptides, mostly self peptides derived from protein turnover and defective ribosomal products. During viral infection, intracellular microorganism infection, or cancerous transformation, such proteins degraded in the proteosome are as well loaded onto MHC class I molecules and displayed on the cell surface. T lymphocytes can detect a peptide displayed at 0.1%-1% of the MHC molecules.

| Characteristic | MHC-I pathway | MHC-II pathway |
|---|---|---|
| Composition of the stable peptide-MHC complex | Polymorphic chain α and β2 microglobulin, peptide bound to α chain | Polymorphic chains α and β, peptide binds to both |
| Types of antigen presenting cells (APC) | All nucleated cells | Dendritic cells, mononuclear phagocytes, B lymphocytes, some endothelial cells, epithelium of thymus |
| T lymphocytes able to respond | Cytotoxic T lymphocytes(CD8+) | Helper T lymphocytes(CD4+) |
| Origin of antigenic proteins | cytosolic proteins (mostly synthetized by the cell; may also enter from the extracellular medium via phagosomes) | Proteins present in endosomes or lysosomes(mostly internalized from extracellular medium) |
| Enzymes responsible for peptide generation | Cytosolic proteasome | Proteases from endosomes and lysosomes (for instance, cathepsin) |
| Location of loading the peptide on the MHC molecule | Endoplasmic reticulum | Specialized vesicular compartment |
| Molecules implicated in transporting the peptides and loading them on the MHC molecules | TAP (transporter associated with antigen processing) | DM, invariant chain |
T lymphocyte recognition restrictions
In their development in the thymus, T lymphocytes are selected to recognize MHC molecules of the host, but not recognize other self antigens. Following selection, each T lymphocyte shows dual specificity: The TCR recognizes self MHC, but only non-self antigens.
MHC restriction occurs during lymphocyte development in the thymus through a process known as positive selection. T cells that do not receive a positive survival signal — mediated mainly by thymic epithelial cells presenting self peptides bound to MHC molecules — to their TCR undergo apoptosis. Positive selection ensures that mature T cells can functionally recognize MHC molecules in the periphery (i.e. elsewhere in the body).
The TCRs of T lymphocytes recognise only sequential epitopes, also called linear epitopes, of only peptides and only if coupled within an MHC molecule. (Antibody molecules secreted by activated B cells, though, ligate diverse epitopes—peptide, lipid, carbohydrate, and nucleic acid—and recognize conformational epitopes, which have three-dimensional structure.)
In sexual mate selection
MHC molecules enable immune system surveillance of the population of protein molecules in a host cell, and greater MHC diversity permits greater diversity of antigen presentation. In 1976, Yamazaki et al demonstrated a sexual selection mate choice by male mice for females of a different MHC. Similar results have been obtained with fish. Some data find lower rates of early pregnancy loss in human couples of dissimilar MHC genes.
MHC may be related to mate choice in some human populations, a theory that found support by studies by Ober and colleagues in 1997, as well as by Chaix and colleagues in 2008.[17] However, the latter findings have been controversial. If it exists, the phenomenon might be mediated by olfaction, as MHC phenotype appears strongly involved in the strength and pleasantness of perceived odour of compounds from sweat. Fatty acid esters—such as methyl undecanoate, methyl decanoate, methyl nonanoate, methyl octanoate, and methyl hexanoate—show strong connection to MHC.
In 1995, Claus Wedekind found that in a group of female college students who smelled T-shirts worn by male students for two nights (without deodorant, cologne, or scented soaps), by far most women chose shirts worn by men of dissimilar MHCs, a preference reversed if the women were on oral contraceptives. Results of a 2002 experiment likewise suggest HLA-associated odors influence odor preference and may mediate social cues. In 2005 in a group of 58 subjects, women were more indecisive when presented with MHCs like their own, although with oral contraceptives, the women showed no particular preference.[23] No studies show the extent to which odor preference determines mate selection (or vice versa).
Evolutionary diversity
Most mammals have MHC variants similar to those of humans, who bear great allelic diversity, especially among the nine classical genes—seemingly due largely to gene duplication—though human MHC regions have many pseudogenes. The most diverse loci, namely HLA-A, HLA-B, and HLA-DRB1, have roughly 1000, 1600, and 870 known alleles, respectively[citation needed]. Many HLA alleles are ancient, sometimes of greater homology to a chimpanzee MHC alleles than to some other human alleles of the same gene.
MHC allelic diversity has challenged evolutionary biologists for explanation. Most posit balancing selection (see polymorphism (biology)), which is any natural selection process whereby no single allele is absolutely most fit, such as frequency-dependent selectionand heterozygote advantage. Recent models suggest a high number of alleles is implausible via heterozygote advantage alone.[citation needed]
Pathogenic coevolution, a counterhypothesis, posits that common alleles are under greatest pathogenic pressure, driving positive selection of uncommon alleles—moving targets, so to say, for pathogens. As pathogenic pressure on the previously common alleles decreases, their frequency in the population stabilizes, and remain circulating in a large population. Despite great MHC polymorphism at the population level, an individual bears at most 18 MHC I or II alleles.
Relatively low MHC diversity has been observed in the cheetah (Acinonyx jubatus),[25]Eurasian beaver (Castor fiber),[26] and giant panda (Ailuropoda melanoleuca).[27] In 2007 low MHC diversity was attributed a role in disease susceptibility in the Tasmanian devil(Sarcophilus harrisii), native to the isolated island of Tasmania, such that an antigen of a transmissible tumor, involved in devil facial tumour disease, appears to be recognized as a self antigen.[28] To offset inbreeding, efforts to sustain genetic diversity in populations of endangered species and of captive animals have been suggested.
In transplant rejection
In a transplant procedure, as of an organ or stem cells, MHC molecules act themselves as antigens and can provoke immune response in the recipient, thus causing transplant rejection. MHC molecules were identified and named after their role in transplantrejection between mice of different strains, though it took over 20 years to clarify MHC’s role in presenting peptide antigens to cytotoxic T lymphocytes (CTLs).
Each human cell expresses six MHC class I alleles (one HLA-A, -B, and -C allele from each parent) and six to eight MHC class II alleles (one HLA-DP and -DQ, and one or two HLA-DR from each parent, and combinations of these). The MHC variation in the human population is high, at least 350 alleles for HLA-A genes, 620 alleles for HLA-B, 400 alleles for DR, and 90 alleles for DQ. Any two individuals who are not identical twins will express differing MHC molecules. All MHC molecules can mediate transplant rejection, but HLA-C and HLA-DP, showing low polymorphism, seem least important.[clarification needed]
When maturing in the thymus, T lymphocytes are selected for their TCR incapacity to recognize self antigens, yet T lymphocytes can react against the donor MHC’s peptide-binding groove, the variable region of MHC holding the presented antigen’s epitope for recognition by TCR, the matching paratope. T lymphocytes of the recipient take the incompatible peptide-binding groove as nonself antigen. The T lymphocytes’ recognition of the foreign MHC as self is allorecognition.
Transplant rejection has various types known to be mediated by MHC (HLA):
- Hyperacute rejection occurs when, before the transplantation, the recipient has preformed anti-HLA antibodies, perhaps by previous blood transfusions (donor tissue that includes lymphocytes expressing HLA molecules), by anti-HLA generated during pregnancy (directed at the father’s HLA displayed by the fetus), or by previous transplantation;
- Acute cellular rejection occurs when the recipient’s T lymphocytes are activated by the donor tissue, causing damage via mechanisms such as direct cytotoxicity from CD8 cells.
- Acute humoral rejection and chronic disfunction occurs when the recipient’s anti-HLA antibodies form directed at HLA molecules present on endothelial cells of the transplanted tissue.
In all of the above situations, immunity is directed at the transplanted organ, sustaining lesions. A cross-reaction test between potential donor cells and recipient serum seeks to detect presence of preformed anti-HLA antibodies in the potential recipient that recognize donor HLA molecules, so as to prevent hyperacute rejection. In normal circumstances, compatibility between HLA-A, -B, and -DR molecules is assessed. The higher the number of incompatibilities, the lower the five-year survival rate. Global databases of donor information enhance the search for compatible donors.
HLA biology

The most studied HLA genes are the nine classical MHC genes: HLA-A, HLA-B, HLA-C, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, and HLA-DRB1. In humans, the MHC gene cluster is divided into three regions: classes I, II, and III. The A, B and C genes belong to MHC class I, whereas the six D genes belong to class II.
MHC alleles are expressed in codominant fashion.[11] This means the alleles (variants) inherited from both parents are expressed equally:
- Each person carries 2 alleles of each of the 3 class-I genes, (HLA-A, HLA-B and HLA-C), and so can express six different types of MHC-I (see figure).
- In the class-II locus, each person inherits a pair of HLA-DP genes (DPA1 and DPB1, which encode α and β chains), a couple of genes HLA-DQ (DQA1 and DQB1, for α and β chains), one gene HLA-DRα (DRA1), and one or more genes HLA-DRβ (DRB1and DRB3, -4 or -5). That means that one heterozygous individual can inherit six or eight functioning class-II alleles, three or more from each parent. The role of DQA2or DQB2 is not verified. The DRB2, DRB6, DRB7, DRB8 and DRB9 are pseudogenes.
The set of alleles that is present in each chromosome is called the MHC haplotype. In humans, each HLA allele is named with a number. For instance, for a given individual, his haplotype might be HLA-A2, HLA-B5, HLA-DR3, etc… Each heterozygous individual will have two MHC haplotypes, one each from the paternal and maternal chromosomes.
The MHC genes are highly polymorphic; many different alleles exist in the different individuals inside a population. The polymorphism is so high, in a mixed population (nonendogamic), no two individuals have exactly the same set of MHC molecules, with the exception of identical twins.
The polymorphic regions in each allele are located in the region for peptide contact. Of all the peptides that could be displayed by MHC, only a subset will bind strongly enough to any given HLA allele, so by carrying two alleles for each gene, a much larger set of peptides can be presented.[why?]
On the other hand, inside a population, the presence of many different alleles ensures there will always be an individual with a specific MHC molecule able to load the correct peptide to recognize a specific microbe. The evolution of the MHC polymorphism ensures that a population will not succumb to a new pathogen or a mutated one, because at least some individuals will be able to develop an adequate immune response to win over the pathogen. The variations in the MHC molecules (responsible for the polymorphism) are the result of the inheritance of different MHC molecules, and they are not induced by recombination, as it is the case for the antigen receptors.
Because of the high levels of allelic diversity found within its genes, MHC has also attracted the attention of many evolutionary biologists.
Immunoglobulin superfamily
The immunoglobulin superfamily (IgSF) is a large group of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins (also known as antibodies); they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. The sperm-specific protein Izumo, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.
Immunoglobulin domains
Proteins of the IgSF possess a structural domain known as an immunoglobulin (Ig) domain. Ig domains are named after the immunoglobulin molecules. They contain about 70-110 amino acids and are categorized according to their size and function. Ig-domains possess a characteristic Ig-fold, which has a sandwich-like structure formed by two sheets of antiparallel beta strands. Interactions between hydrophobic amino acids on the inner side of the sandwich and highly conserved disulfide bonds formed between cysteine residues in the B and F strands, stabilize the Ig-fold. One end of the Ig domain has a section called the complementarity determining region that is important for the specificity of antibodies for their ligands.
Classification
The Ig like domains can be classified as IgV, IgC1, IgC2, or IgI.
Most Ig domains are either variable (IgV) or constant (IgC).
IgV: IgV domains with 9 beta strands are generally longer than IgC domains with 7 beta strands.
IgC1 and IgC2: Ig domains of some members of the IgSF resemble IgV domains in the amino acid sequence, yet are similar in size to IgC domains. These are called IgC2 domains, while standard IgC domains are called IgC1 domains.
IgI: Other Ig domains exist that are called intermediate (I) domains.
Members of the immunoglobulin superfamily
The Ig domain was reported to be the most populous family of proteins in the human genome with 765 members identified. Members of the family can be found even in the bodies of animals with a simple physiological structure such as poriferan sponges. They have also been found in bacteria, where their presence is thought to be due to horizontal gene transfer.
| Molecule function/category | Examples | Description |
|---|---|---|
| Antigen receptors |
|
Antigen receptors found on the surface of T and B lymphocytes in all jawed vertebrates belong to the IgSF. Immunoglobulin molecules (the antigen receptors of B cells) are the founding members of the IgSF. In humans, there are five distinct types of immunoglobulinmolecule all containing a heavy chain with four Ig domains and a light chain with two Ig domains. The antigen receptor of T cells is the T cell receptor (TCR), which is composed of two chains, either the TCR-alpha and -beta chains, or the TCR-delta and gamma chains. All TCR chains contain two Ig domains in the extracellular portion; one IgV domain at the N-terminus and one IgC1 domain adjacent to the cell membrane. |
| Antigen presentingmolecules |
|
The ligands for TCRs are major histocompatibility complex (MHC) proteins. These come in two forms; MHC class I forms a dimer with a molecule called beta-2 microglobulin (β2M) and interacts with the TCR on cytotoxic T cells and MHC class II has two chains (alpha and beta) that interact with the TCR on helper T cells. MHC class I, MHC class II and β2M molecules all possess Ig domains and are therefore also members of the IgSF. |
| Co-receptors |
|
Co-receptors and accessory molecules: Other molecules on the surfaces of T cells also interact with MHC molecules during TCR engagement. These are known as co-receptors. In lymphocyte populations, the co-receptor CD4is found on helper T cells and the co-receptor CD8 is found on cytotoxic T cells. CD4 has four Ig domains in its extracellular portion and functions as a monomer. CD8, in contrast, functions as a dimer with either two identical alpha chains or, more typically, with an alpha and beta chain. CD8-alpha and CD8-beta each has one extracellular IgV domain in its extracellular portion. A co-receptor complex is also used by the BCR, including CD19, an IgSF molecule with two IgC2-domains. |
| Antigen receptor accessory molecules |
|
A further molecule is found on the surface of T cells that is also involved in signaling from the TCR. CD3 is a molecule that helps to transmit a signal from the TCR following its interaction with MHC molecules. Three different chains make up CD3 in humans, the gamma chain, delta chain and epsilon chain, all of which are IgSF molecules with a single Ig domain.Similar to the situation with T cells, B cells also have cell surface co-receptors and accessory molecules that assist with cell activation by the B Cell Receptor (BCR)/immunoglobulin. Two chains are used or signaling, CD79a and CD79b that both possess a single Ig domain. |
| Co-stimulatory or inhibitory molecules |
|
Co-stimulatory or inhibitory molecules: Co-stimulatory and inhibitory signaling receptors and ligands control the activation, expansion and effector functions of cells. One major group of IgSF co-stimulatory receptors are molecules of the CD28 family; CD28, CTLA-4, program death-1 (PD-1), the B- and T-lymphocyte attenuator (BTLA, CD272), and the inducible T-cell co-stimulator (ICOS, CD278);[7] and their IgSF ligands belong to the B7 family; CD80 (B7-1), CD86 (B7-2), ICOS ligand, PD-L1 (B7-H1), PD-L2 (B7-DC), B7-H3, and B7-H4 (B7x/B7-S1).[8] |
| Receptors on Natural killer cells |
|
|
| Receptors on Leukocytes |
|
|
| IgSF CAMs |
|
|
| Cytokine receptors |
|
|
| Growth factor receptors |
|
|
| Receptor tyrosine kinases/phosphatases |
|
|
| Ig binding receptors |
|
|
| Others |
|
MHC class I
MHC class I molecules are one of two primary classes of major histocompatibility complex (MHC) molecules (the other being MHC class II) and are found on the cell surface of all nucleated cells in the bodies of jawed vertebrates. They also occur on platelets, but not on red blood cells. Their function is to display peptide fragments of non-self proteins from within the cell to cytotoxic T cells; this will trigger an immediate response from the immune system against a particular non-self antigen displayed with the help of an MHC class I protein. Because MHC class I molecules present peptides derived from cytosolic proteins, the pathway of MHC class I presentation is often called cytosolic or endogenous pathway.
Function
Class I MHC molecules bind peptides generated mainly from degradation of cytosolic proteins by the proteasome. The MHC I:peptide complex is then inserted via endoplasmic reticulum into the external plasma membrane of the cell. The epitope peptide is bound on extracellular parts of the class I MHC molecule. Thus, the function of the class I MHC is to display intracellular proteins to cytotoxic T cells (CTLs). However, class I MHC can also present peptides generated from exogenous proteins, in a process known as cross-presentation.
A normal cell will display peptides from normal cellular protein turnover on its class I MHC, and CTLs will not be activated in response to them due to central and peripheral tolerance mechanisms. When a cell expresses foreign proteins, such as after viral infection, a fraction of the class I MHC will display these peptides on the cell surface. Consequently, CTLs specific for the MHC:peptide complex will recognize and kill presenting cells.
Alternatively, class I MHC itself can serve as an inhibitory ligand for natural killer cells (NKs). Reduction in the normal levels of surface class I MHC, a mechanism employed by some viruses during immune evasion or in certain tumors, will activate NK cell killing.
Locus
A locus (plural loci) in genetics is the position of a gene on a chromosome. Each chromosome carries many genes; humans’ estimated ‘haploid’ protein coding genes are 19,000-20,000,[2] on the 23 different chromosomes. A variant of the similar DNA sequence located at a given locus is called an allele. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the locus for a particular biological trait.
Diploid and polyploid cells whose chromosomes have the same allele of a given gene at some locus are called homozygous with respect to that gene, while those that have different alleles of a given gene at a locus, are called heterozygous with respect to that gene.
Nomenclature
The chromosomal locus of a gene might be written “6p21.3”. Because “21” refers to “region 2, band 1” this is read as “two one”, not as “twenty-one”. So the entire locus is “six P two one point three.”
| Component | Explanation |
| 6 | The chromosome number. |
| p | The position is on the chromosome’s short arm (a common apocryphal explanation is that the p stands for petit in French); qindicates the long arm (chosen as next letter in alphabet after p; alternatively it is sometimes said that q stands for queuemeaning tail in French). |
| 21.3 | The numbers that follow the letter represent the position on the arm: region 2, band 1, sub-band 3. The bands are visible under a microscope when chromosome is suitably stained. Each of the bands is numbered, beginning with 1 for the band nearest the centromere. Sub-bands and sub-sub-bands are visible at higher resolution. |
A range of loci is specified in a similar way. For example, the locus of gene OCA1 may be written “11q1.4-q2.1”, meaning it is on the long arm of chromosome 11, somewhere in the range from sub-band 4 of region 1 to sub-band 1 of region 2.
The ends of a chromosome are labeled “pter” and “qter”, and so “2qter” refers to the terminus of the long arm of chromosome 2.
Centisome
A centisome (not to be confused with a centrosome) is defined as 1% of a chromosome length.
Liste des allèles humains d’antigène leucocytaire associés aux affections cutanées
Il existe de nombreux allèles de l’antigène leucocytaire humain (HLA) associés à des affections ou affectant le système tégumentaire humain
| Condition | Associated HLA allele(s) |
|---|---|
| Fixed drug eruption | B22 |
| Lichen planus | DR1 DR2 DRw9 DR10 Bw15 B8 |
| Psoriasis | Cw6 DR406 |
| Psoriatic arthritis | B27 |
| Ankylosing spondylitis | B27 |
| Reactive arthritis | B27 |
| Acute anterior uveitis | B27 |
| Behçet’s disease | B51 |
| Dermatitis herpetiformis | DQw2 DR3 B8 |
| Pemphigus vulgaris | DR4 DRw6 Dw10 |
| Herpes gestationis | DR3 DR4 |
| Epidermolysis bullosa acquisita | DR2 |
| Subacute cutaneous lupus erythematosus | DR3 |
| Dermatomyositis | DQA1 DR3 B8 DRw52 |
| Alopecia areata | DR5 |
| Sjögren’s syndrome | DR3 DQ2 |
| Herpes simplex virus-related erythema multiforme | B12 |
| Chronic idiopathic urticaria | DR4 DQ8 |
| Actinic prurigo | DR4 subtype DRB1*0407 |
| Systemic lupus erythematosus | DR3 DR2 |
| Generalized granuloma annulare | Bw35 |
| Lichen sclerosus | DQ7 |
| Early onset and severe psoriasis | B17 |
